2A4G
 
 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5J2V
 
 | | Crystal Structure of Hsp90-alpha Apo N-domain | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
4W78
 
 | | Crystal structure of the ChsH1-ChsH2 complex from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-08-21 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | A Distinct MaoC-like Enoyl-CoA Hydratase Architecture Mediates Cholesterol Catabolism in Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
2Q0S
 
 | | Structure of the Inhibitor bound form of M. Smegmatis Aryl Esterase | | Descriptor: | Aryl esterase, SULFATE ION | | Authors: | Mathews, I.I, Soltis, M, Saldajeno, M, Ganshaw, G, Sala, R, Weyler, W, Cervin, M.A, Whited, G, Bott, R. | | Deposit date: | 2007-05-22 | | Release date: | 2007-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a novel enzyme that catalyzes acyl transfer to alcohols in aqueous conditions.
Biochemistry, 46, 2007
|
|
5J64
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with 5-(2,4-Dihydroxy-phenyl)-4-(2-fluoro-phenyl)-2,4-dihydro-[1,2,4]triazol-3-one | | Descriptor: | 5-(2,4-dihydroxyphenyl)-4-(2-fluorophenyl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
5J20
 
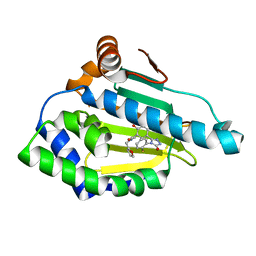 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-N-furan-2-ylmethyl-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(furan-2-yl)methyl]-2,4-dihydroxy-N-methylbenzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
9AX6
 
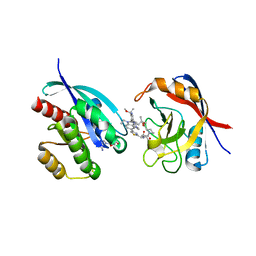 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
5J72
 
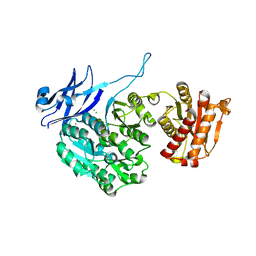 | | Cwp6 from Clostridium difficile | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
1GX3
 
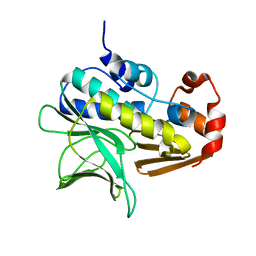 | | M. smegmatis arylamine N-acetyl transferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Mushtaq, A, Kawamura, A, Sinclair, J, Sim, E, Noble, M. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Arylamine N-Acetyltransferase from Mycobacterium Smegmatis-an Enzyme which Inactivates the Anti-Tubercular Drug, Isoniazid
J.Mol.Biol., 318, 2002
|
|
7X91
 
 | |
3KB1
 
 | | Crystal Structure of the Nucleotide-binding protein AF_226 in complex with ADP from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR157 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nucleotide-binding protein, ZINC ION | | Authors: | Forouhar, F, Lew, S, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR157
To be Published
|
|
1QF3
 
 | | PEANUT LECTIN COMPLEXED WITH METHYL-BETA-GALACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6Z35
 
 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
7X8W
 
 | |
3KLU
 
 | | Crystal structure of the protein yqbn. northeast structural genomics consortium target sr445. | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein yqbN | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, M, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the protein yqbn. northeast structural genomics consortium target sr445.
To be Published
|
|
5J6L
 
 | | Crystal Structure of Hsp90-alpha N-domain in complex with N-Butyl-5-[4-(2-fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-benzamide | | Descriptor: | Heat shock protein HSP 90-alpha, N-butyl-5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methylbenzamide | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
1QUE
 
 | | X-RAY STRUCTURE OF THE FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 1.8 ANGSTROMS | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-07-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
7X8Z
 
 | |
1QUF
 
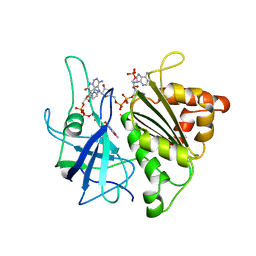 | | X-RAY STRUCTURE OF A COMPLEX NADP+-FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 2.25 ANGSTROMS | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-09-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
7X8Y
 
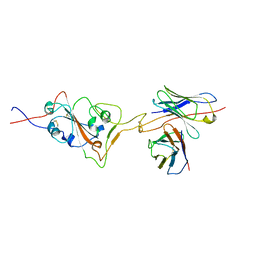 | |
5J86
 
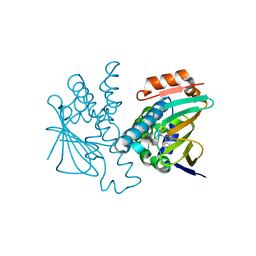 | | Crystal Structure of Hsp90-alpha N-domain in complex with 2,4-Dihydroxy-N-methyl-5-(5-oxo-4-o-tolyl-4,5-dihydro-1H-[1,2,4]triazol-3-yl)-N-thiophen-2-ylmethyl-benzamide | | Descriptor: | 2,4-dihydroxy-N-methyl-5-[4-(2-methylphenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-N-[(thiophen-2-yl)methyl]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
7X90
 
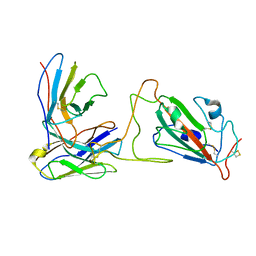 | |
7X92
 
 | |
5JHR
 
 | | Yeast 20S proteasome in complex with the peptidic epoxyketone inhibitor 27 | | Descriptor: | (2S)-2-azido-N-[(2S)-3-(biphenyl-4-yl)-1-{[(2S)-1-{[(2S,3S,4R)-3,5-dihydroxy-4-methylpentan-2-yl]amino}-1-oxo-3-phenylpropan-2-yl]amino}-1-oxopropan-2-yl]-3-phenylpropanamide (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2016-04-21 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of beta 5c Selective Inhibitors of Human Constitutive Proteasomes.
J.Med.Chem., 59, 2016
|
|
