4X29
 
 | |
2E7K
 
 | | Solution structure of the PDZ domain from Human MAGUK p55 subfamily member 2 | | Descriptor: | MAGUK p55 subfamily member 2 | | Authors: | Qin, X.R, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-10 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain from Human MAGUK p55 subfamily member 2
To be Published
|
|
7P5Q
 
 | |
1A3Y
 
 | |
2DY4
 
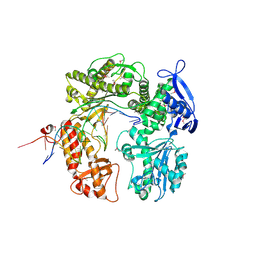 | | Crystal structure of RB69 GP43 in complex with DNA containing Thymine Glycol | | Descriptor: | 5'-D(*CP*GP*(CTG)P*GP*GP*AP*AP*TP*GP*A*CP*AP*GP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*CP*TP*GP*T*CP*AP*TP*TP*CP*CP*A)-3', DNA polymerase | | Authors: | Aller, P, Rould, M.A, Hogg, M, Wallace, S.S, Doublie, S. | | Deposit date: | 2006-09-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2E7V
 
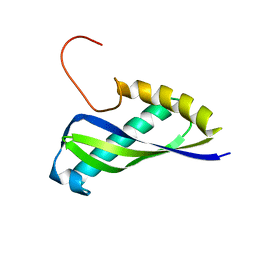 | | Crystal structure of SEA domain of transmembrane protease from Mus musculus | | Descriptor: | Transmembrane protease | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Shirozu, M, Yokoyama, S, Chen, L, Liu, Z.J, Wang, B.C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SEA domain of transmembrane protease from Mus musculus
To be Published
|
|
2E89
 
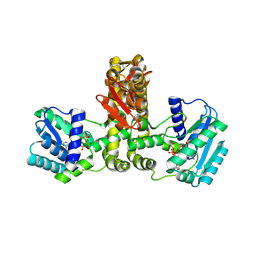 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
2EAC
 
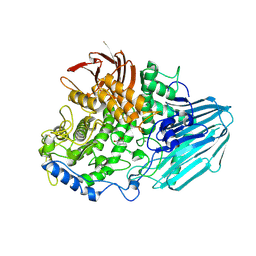 | | Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, Alpha-fucosidase, CALCIUM ION | | Authors: | Nagae, M, Tsuchiya, A, Katayama, T, Yamamoto, K, Wakatsuki, S, Kato, R. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis on the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase (AFCA) from Bifidobacterium bifidum
J.Biol.Chem., 282, 2007
|
|
7P5S
 
 | |
7P5G
 
 | |
2E25
 
 | | The Crystal Structure of the T109S mutant of E. coli Dihydroorotase complexed with an inhibitor 5-fluoroorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2006-11-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the T109S mutant of Escherichia coli dihydroorotase complexed with the inhibitor 5-fluoroorotate: catalytic activity is reflected by the crystal form
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2EDV
 
 | | Solution structure of the PDZ domain from human FERM and PDZ domain containing 1 | | Descriptor: | FERM and PDZ domain-containing protein 1 | | Authors: | Seimiya, K, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain from human FERM and PDZ domain containing 1
to be published
|
|
2E2T
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with phenylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E3S
 
 | | Crystal structure of CERT START domain co-crystallized with C24-ceramide (P21) | | Descriptor: | Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2006-11-28 | | Release date: | 2007-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2EJ8
 
 | | Crystal structure of APPL1 PTB domain at 1.8A | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Tanabe, H, Hosaka, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2008-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of APPL1 PTB domain at 1.8A
To be Published
|
|
2EJN
 
 | | Structural characterization of the tetrameric form of the major cat allergen fel D 1 | | Descriptor: | CALCIUM ION, Major allergen I polypeptide chain 1, chain 2 | | Authors: | Kaiser, L, Velickovic, T.C, Badia-Martinez, D, Adedoyin, J, Thunberg, S, Hallen, D, Berndt, K, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the tetrameric form of the major cat allergen Fel d 1
J.Mol.Biol., 370, 2007
|
|
2EL1
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L44M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L44M)
To be Published
|
|
2ELN
 
 | | Solution structure of the 11th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 11th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
2ELT
 
 | | Solution structure of the 3rd C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 3rd C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
2EM4
 
 | | Solution structure of the C2H2 type zinc finger (region 724-756) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 724-756) of human Zinc finger protein 28 homolog
To be Published
|
|
2E63
 
 | | Solution structure of the NEUZ domain in KIAA1787 protein | | Descriptor: | KIAA1787 protein | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
2EMJ
 
 | | Solution structure of the C2H2 type zinc finger (region 612-644) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 612-644) of human Zinc finger protein 28 homolog
To be Published
|
|
2EN0
 
 | | Solution structure of the C2H2 type zinc finger (region 385-413) of human Zinc finger protein 268 | | Descriptor: | ZINC ION, Zinc finger protein 268 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 385-413) of human Zinc finger protein 268
To be Published
|
|
2ENF
 
 | | Solution structure of the C2H2 type zinc finger (region 340-372) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 340-372) of human Zinc finger protein 347
To be Published
|
|
2E2U
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with 4-hydroxybenzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
