4AB7
 
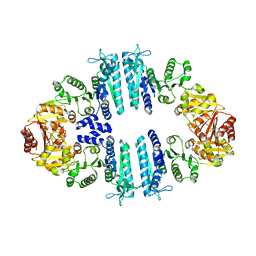 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae complexed with its substrate N- acetylglutamate | | Descriptor: | N-ACETYL-L-GLUTAMATE, PROTEIN ARG5,6, MITOCHONDRIAL | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
2MW2
 
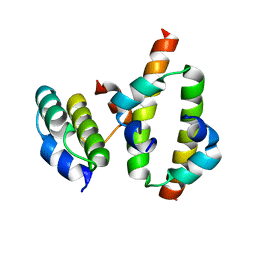 | | Hha-H-NS46 charge zipper complex | | Descriptor: | DNA-binding protein H-NS, Hemolysin expression-modulating protein Hha | | Authors: | Cordeiro, T.N, Garcia, J, Bernado, P, Millet, O, Pons, M. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Three-protein Charge Zipper Stabilizes a Complex Modulating Bacterial Gene Silencing.
J. Biol. Chem., 290, 2015
|
|
4AOF
 
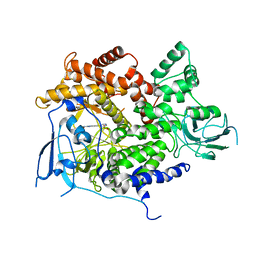 | | Selective small molecule inhibitor discovered by chemoproteomic assay platform reveals regulation of Th17 cell differentiation by PI3Kgamma | | Descriptor: | N-[6-(5-methylsulfonylpyridin-3-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]ethanamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Bergamini, G, Bell, K, Shimamura, S, Werner, T, Cansfield, A, Muller, K, Perrin, J, Rau, C, Ellard, K, Hopf, C, Doce, C, Leggate, D, Mangano, R, Mathieson, T, OMahony, A, Plavec, I, Rharbaoui, F, Reinhard, F, Savitski, M.M, Ramsden, N, Hirsch, E, Drewes, G, Rausch, O, Bantscheff, M, Neubauer, G. | | Deposit date: | 2012-03-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Selective Inhibitor Reveals Pi3Kgamma Dependence of T(H)17 Cell Differentiation.
Nat.Chem.Biol., 8, 2012
|
|
4A0V
 
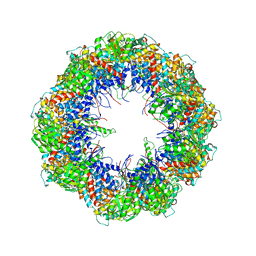 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
4A1Y
 
 | | Human myelin P2 protein, K65Q mutant | | Descriptor: | CHLORIDE ION, MYELIN P2 PROTEIN, PALMITIC ACID | | Authors: | Lehtimaki, M, Kursula, P. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Function Relationships in the Myelin Peripheral Membrane Protein P2
To be Published
|
|
2MPV
 
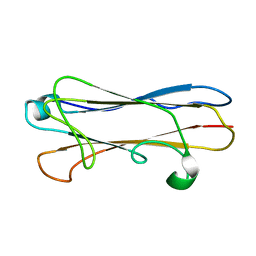 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
4A4Z
 
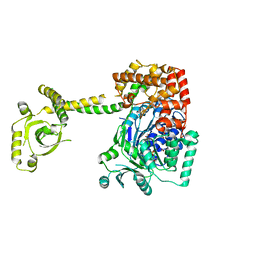 | | CRYSTAL STRUCTURE OF THE S. CEREVISIAE DEXH HELICASE SKI2 BOUND TO AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, ANTIVIRAL HELICASE SKI2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Halbach, F, Rode, M, Conti, E. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of S. Cerevisiae Ski2, a Dexh Helicase Associated with the Cytoplasmic Functions of the Exosome.
RNA, 18, 2012
|
|
2MQB
 
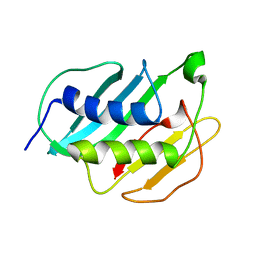 | |
4AB3
 
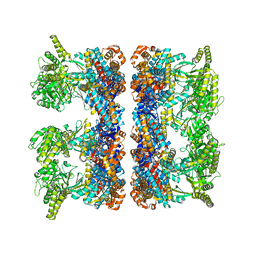 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
2N2G
 
 | |
4ZDV
 
 | |
4ADX
 
 | | The Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 | | Descriptor: | 23S Ribosomal RNA EXPANSION SEGMENTS, 23S ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Greber, B.J, Boehringer, D, Godinic-Mikulcic, V, Crnkovic, A, Ibba, M, Weygand-Durasevic, I, Ban, N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-Em Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution
J.Mol.Biol., 418, 2012
|
|
2N0F
 
 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2QH5
 
 | | Crystal structure of mannose-6-phosphate isomerase from Helicobacter pylori | | Descriptor: | Mannose-6-phosphate isomerase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Iizuka, M, Maletic, M, Wasserman, S.R, Koss, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mannose-6-phosphate isomerase from Helicobacter pylori.
To be Published
|
|
2N79
 
 | | The structural and functional effects of the Familial Hypertrophic Cardiomyopathy-linked cardiac troponin C mutation, L29Q | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Sevrieva, I, Li, M.X, Irving, M, Sun, Y, Sykes, B. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural and functional effects of the familial hypertrophic cardiomyopathy-linked cardiac troponin C mutation, L29Q.
J.MOL.CELL.CARDIOL., 87, 2015
|
|
4AK1
 
 | | Structure of BT4661, a SusE-like surface located polysaccharide binding protein from the Bacteroides thetaiotaomicron heparin utilisation locus | | Descriptor: | BT_4661, SODIUM ION | | Authors: | Lowe, E.C, Basle, A, Czjzek, M, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2012-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2MWM
 
 | |
4AKT
 
 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AGD
 
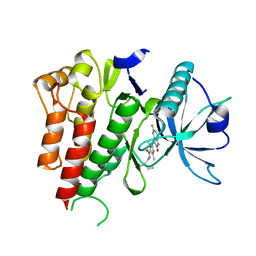 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH SUNITINIB (SU11248) (N-2-diethylaminoethyl)-5-((Z)-(5- fluoro-2-oxo-1H-indol-3-ylidene)methyl)-2,4-dimethyl-1H-pyrrole-3- carboxamide) | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | McTigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Stewart, A. | | Deposit date: | 2012-01-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AR2
 
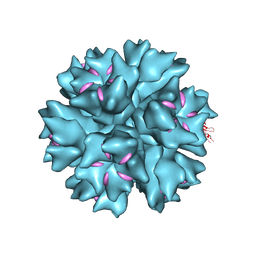 | | Dodecahedron formed of penton base protein from adenovirus Ad3 | | Descriptor: | CALCIUM ION, FIBER PROTEIN, L2 PROTEIN III (PENTON BASE) | | Authors: | Burmeister, W.P, Szolajska, E, Zochowska, M, Nerlo, B, Andreev, I, Schoehn, G, Andrieu, J.-P, Fender, P, Naskalska, A, Zubieta, C, Cusack, S, Chroboczek, J. | | Deposit date: | 2012-04-20 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis for the Integrity of Adenovirus Ad3 Dodecahedron.
Plos One, 7, 2012
|
|
4F11
 
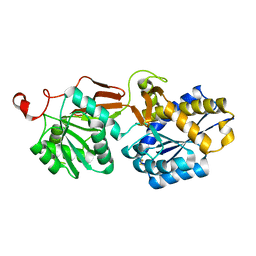 | | Crystal structure of the extracellular domain of human GABA(B) receptor GBR2 | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Geng, Y, Xiong, D, Mosyak, L, Malito, D.L, Kniazeff, J, Chen, Y, Burmakina, S, Quick, M, Bush, M, Javitch, J.A, Pin, J.-P, Fan, Q.R. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and functional interaction of the extracellular domain of human GABA(B) receptor GBR2.
Nat.Neurosci., 15, 2012
|
|
2JIN
 
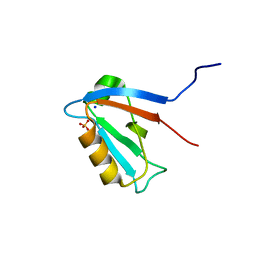 | | Crystal structure of PDZ domain of Synaptojanin-2 binding protein | | Descriptor: | SODIUM ION, SULFATE ION, SYNAPTOJANIN-2 BINDING PROTEIN | | Authors: | Tickle, J, Phillips, C, Pike, A.C.W, Cooper, C, Salah, E, Elkins, J, Turnbull, A.P, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Pdz Domain of Synaptojanin-2 Binding Protein
To be Published
|
|
4AQG
 
 | | X-ray crystallographic structure of Crimean-congo haemorrhagic fever virus nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Wang, Y, Dutta, S, Karlberg, H, Devignot, S, Weber, F, Hao, Q, Tan, Y.J, Mirazimi, A, Kotaka, M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Crimean-Congo Haemorraghic Fever Virus Nucleoprotein: Superhelical Homo-Oligomers and the Role of Caspase-3 Cleavage.
J.Virol., 86, 2012
|
|
4AR4
 
 | | Neutron crystallographic structure of the reduced form perdeuterated Pyrococcus furiosus rubredoxin to 1.38 Angstrom resolution. | | Descriptor: | FE (III) ION, Rubredoxin, deuterium(1+), ... | | Authors: | Cuypers, M.G, Mason, S.A, Blakeley, M.P, Mitchell, E.P, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2012-04-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-06-19 | | Method: | NEUTRON DIFFRACTION (1.381 Å) | | Cite: | Near-Atomic Resolution Neutron Crystallography on Perdeuterated Pyrococcus Furiosus Rubredoxin: Implication of Hydronium Ions and Protonation State Equilibria in Redox Changes.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
