5VAI
 
 | | Cryo-EM structure of the activated Glucagon-like peptide-1 receptor in complex with G protein | | Descriptor: | Glucagon-like peptide 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, Y, Sun, B, Feng, D, Hu, H, Chu, M, Qu, Q, Tarrasch, J.T, Li, S, Kobilka, T.S, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2017-03-27 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein.
Nature, 546, 2017
|
|
1KFW
 
 | |
1TRY
 
 | | STRUCTURE OF INHIBITED TRYPSIN FROM FUSARIUM OXYSPORUM AT 1.55 ANGSTROMS | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHORYLISOPROPANE, TRYPSIN | | Authors: | Rypniewski, W.R, Dambmann, C, Von Der Osten, C, Dauter, M, Wilson, K.S. | | Deposit date: | 1994-03-07 | | Release date: | 1996-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of inhibited trypsin from Fusarium oxysporum at 1.55 A.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1KJP
 
 | |
1KGL
 
 | | Solution structure of cellular retinol binding protein type-I in complex with all-trans-retinol | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I, RETINOL | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-11-27 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Backbone Dynamics of Apo- and Holo-cellular Retinol-binding
Protein in Solution.
J.Biol.Chem., 277, 2002
|
|
7FJH
 
 | | LecA from Pseudomonas aeruginosa in complex with 4-Phenylbutyryl hydroxamic acid (CAS: 32153-46-1) | | Descriptor: | CALCIUM ION, N-oxidanyl-4-phenyl-butanamide, PA-I galactophilic lectin | | Authors: | Shanina, S, Kuhaudomlarp, S, Siebs, E, Fuchsberger, F, Denis, M, da Silva Figueiredo Celstino Gomes, P, Clausen, M.H, Seeberger, P.H, Rognan, D, Titz, A, Imberty, A, Rademacher, C. | | Deposit date: | 2021-08-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Targeting undruggable carbohydrate recognition sites through focused fragment library design.
Commun Chem, 5, 2022
|
|
1KK6
 
 | | Crystal Structure of Vat(D) (Form I) | | Descriptor: | STREPTOGRAMIN A ACETYLTRANSFERASE | | Authors: | Sugantino, M, Roderick, S.L. | | Deposit date: | 2001-12-06 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics.
Biochemistry, 41, 2002
|
|
5V2V
 
 | | Ethylene forming enzyme in complex with nickel | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, NICKEL (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5V2Y
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and L-arginine | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2XPL
 
 | | Crystal structure of Iws1(Spn1) conserved domain from Encephalitozoon cuniculi | | Descriptor: | CHLORIDE ION, IWS1 | | Authors: | Koch, M, Diebold, M.-L, Cura, V, Cavarelli, J, Romier, C. | | Deposit date: | 2010-08-27 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Iws1/Spt6 Complex Reveals an Interaction Domain Conserved in Tfiis, Elongin a and Med26
Embo J., 29, 2010
|
|
5UTO
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, Broussard, T.C, Miller, D.J, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-15 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
7F4B
 
 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KQQ
 
 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
7FFM
 
 | | Human serum transferrin with five osmium binding sites | | Descriptor: | MALONATE ION, NITRILOTRIACETIC ACID, OSMIUM ION, ... | | Authors: | Wang, M, Sun, H. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
2XD5
 
 | | Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b | | Descriptor: | CHLORIDE ION, N-BENZOYL-D-ALANINE, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Lemaire, D, Jamin, M, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism and the Role of Streptococcus Pneumoniae Pbp1B
To be Published
|
|
1KN2
 
 | | CATALYTIC ANTIBODY D2.3 COMPLEX | | Descriptor: | IG ANTIBODY D2.3 (HEAVY CHAIN), IG ANTIBODY D2.3 (LIGHT CHAIN), PARA-NITROPHENYL PHOSPHONOBUTANOYL L-ALANINE, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remarkable remote chiral recognition in a reaction mediated by a catalytic antibody.
J.Am.Chem.Soc., 124, 2002
|
|
5UZ8
 
 | | Crystal Structure of Mouse Cadherin-23 EC22-24 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Patel, A, Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
1KW4
 
 | |
7F4Z
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | Authors: | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
2XKR
 
 | | Crystal Structure of Mycobacterium tuberculosis CYP142: A novel cholesterol oxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 142, TETRAETHYLENE GLYCOL | | Authors: | Driscoll, M, McLean, K.J, Levy, C.W, Lafite, P, Mast, N, Pikuleva, I.A, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium Tuberculosis Cyp142: Evidence for Multiple Cholesterol 27-Hydroxylase Activities in a Human Pathogen.
J.Biol.Chem., 285, 2010
|
|
2XT6
 
 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
5V11
 
 | |
7F4C
 
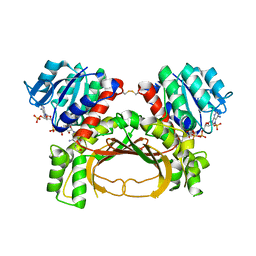 | | The crystal structure of the immature holo-enzyme of homoserine dehydrogenase complexed with NADP and 1,4-butandiol from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | 1,4-BUTANEDIOL, Homoserine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ogata, K, Kaneko, R, Kubota, T, Watanabe, K, Kurihara, E, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F47
 
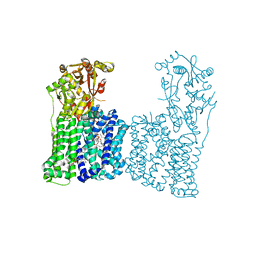 | | Cryo-EM structure of Rhizobium etli MprF | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Hypothetical conserved protein, [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate | | Authors: | Nishimura, M, Hirano, H, Kobayashi, K, Gill, C.P, Phan, C.N.K, Kise, Y, Kusakizako, T, Yamashita, K, Ito, Y, Roy, H, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Rhizobium etli MprF
To Be Published
|
|
