8P7J
 
 | | Crystal structure of MAP2K6 with a covalent compound GCL96 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, N-[3-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]prop-2-enamide | | Authors: | Wang, G.Q, Seidler, N, Roehm, S, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of MAP2K6 with a covalent compound GCL96
To Be Published
|
|
1JBE
 
 | |
6Y83
 
 | | Capsid structure of Leishmania RNA virus 1 | | Descriptor: | Capsid protein | | Authors: | Prochazkova, M, Fuzik, T, Grybtchuk, D, Falginella, F, Podesvova, L, Yurchenko, V, Vacha, R, Plevka, P. | | Deposit date: | 2020-03-03 | | Release date: | 2020-11-11 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Capsid Structure of Leishmania RNA Virus 1.
J.Virol., 95, 2021
|
|
5L65
 
 | |
4NGO
 
 | | Previously de-ionized HEW lysozyme batch crystallized in 1.0 M CoCl2 | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7RE4
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
4NGW
 
 | | Dialyzed HEW lysozyme batch crystallized in 0.5 M YbCl3 and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, YTTERBIUM (III) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6Y9G
 
 | |
6FQA
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Parilova, O, Pakharukova, N.A, Malmi, H, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
7REA
 
 | | Apo Hemophilin from A. baumannii | | Descriptor: | Hemophilin | | Authors: | Bateman, T.J, Shah, M, Moraes, T.F. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Slam-dependent hemophore contributes to heme acquisition in the bacterial pathogen Acinetobacter baumannii.
Nat Commun, 12, 2021
|
|
6Y8C
 
 | |
6YA2
 
 | | Crystal structure of TSWV glycoprotein N ectodomain (Trypsin treated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein | | Authors: | Dessau, M, Bahat, Y. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tomato spotted wilt virus G N reveals a dimer complex formation and evolutionary link to animal-infecting viruses
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1J9V
 
 | | Solution structure of a lactam analogue (DabD) of HIV gp41 600-612 loop. | | Descriptor: | DabD (Ace)IWG(DAB)SGKLIDTTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-05-29 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
5L7K
 
 | |
3TSU
 
 | | Crystal structure of E. coli HypF with AMP-PNP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
2YM0
 
 | | Truncated SipD from Salmonella typhimurium | | Descriptor: | CELL INVASION PROTEIN SIPD, GLYCEROL | | Authors: | Lunelli, M, Kolbe, M. | | Deposit date: | 2011-06-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands.
PLoS Pathog., 7, 2011
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
5EL7
 
 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the second position and antibiotic paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
5L7P
 
 | | In silico-powered specific incorporation of photocaged Dopa at multiple protein sites | | Descriptor: | (2~{S})-2-azanyl-3-[3-[(2-nitrophenyl)methoxy]-4-oxidanyl-phenyl]propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hauf, M, Richter, F, Schneider, T, Martins, B.M, Baumann, T, Durkin, P, Dobbek, H, Moeglich, A, Budisa, N. | | Deposit date: | 2016-06-03 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code.
Chembiochem, 18, 2017
|
|
6Y9K
 
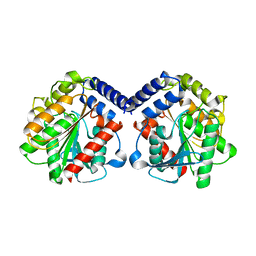 | |
2YMB
 
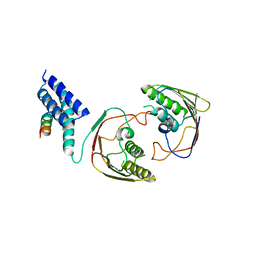 | | Structures of MITD1 | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YNG
 
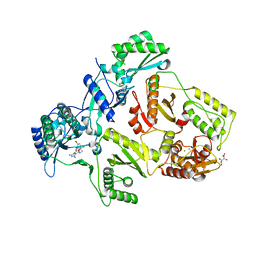 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YPH
 
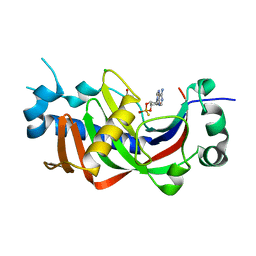 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230S, crystallized with 2',3-(RP)- cyclic-AMPS | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, ACETATE ION, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
8P6K
 
 | |
2YP6
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
