6BAS
 
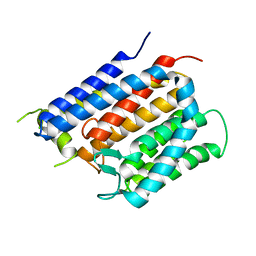 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
3MM0
 
 | | Crystal structure of chimeric avidin | | Descriptor: | Avidin, Avidin-related protein 4/5 | | Authors: | Livnah, O, Eisenberg-Domovich, Y, Maatta, J.A.E, Kulomaa, M.S, Hytonen, V.P, Nordlund, H.R. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chimeric avidin shows stability against harsh chemical conditions-biochemical analysis and 3D structure.
Biotechnol.Bioeng., 108, 2011
|
|
5UG2
 
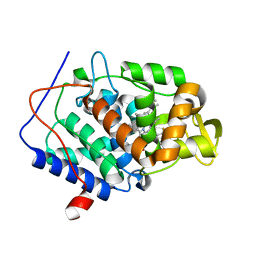 | | CcP gateless cavity | | Descriptor: | 6-fluoro-2-methylimidazo[1,2-a]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Stein, R.M, Fischer, M, Shoichet, B.K. | | Deposit date: | 2017-01-06 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BCK
 
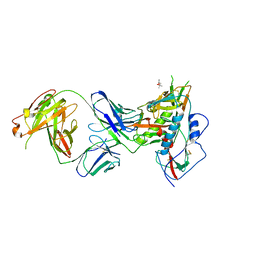 | | Crystal Structure of Broadly Neutralizing Antibody N49P7 in Complex with HIV-1 Clade AE strain 93TH057 gp120 core. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N49P7 Fab heavy chain of N29P7 IgG, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses.
Cell, 173, 2018
|
|
2N3A
 
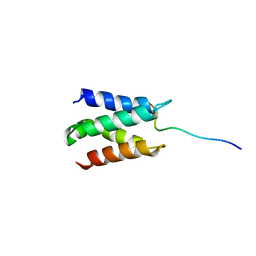 | | Solution structure of LEDGF/p75 IBD in complex with POGZ peptide (1389-1404) | | Descriptor: | PC4 and SFRS1-interacting protein, Pogo transposable element with ZNF domain | | Authors: | Tesina, P, Cermakova, K, Horejsi, M, Prochazkova, K, Fabry, M, Sharma, S, Christ, F, Demeulemeester, J, Debyser, Z, De Rijck, J, Veverka, V, Rezacova, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif.
Nat Commun, 6, 2015
|
|
2MUZ
 
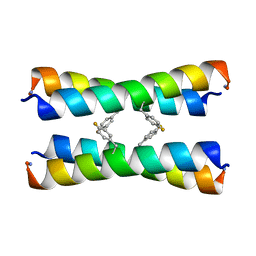 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
2MV4
 
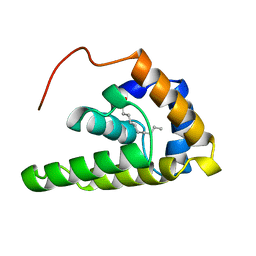 | |
1YOE
 
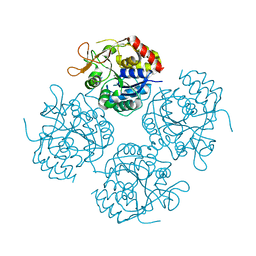 | |
7ZY0
 
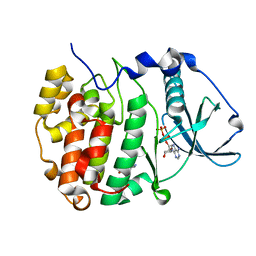 | | Crystal structure of compound 7 bound to CK2alpha | | Descriptor: | 2-(5-bromanyl-1~{H}-indol-3-yl)ethanenitrile, ADENOSINE-5'-DIPHOSPHATE, Casein kinase II subunit alpha | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
7ZY8
 
 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 3-[3,5-bis(chloranyl)phenyl]propan-1-amine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-24 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
6BGJ
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BI2
 
 | |
7ZY5
 
 | | Crystal structure of compound 2 bound to CK2alpha | | Descriptor: | 1-NAPHTHOL, ACETATE ION, Casein kinase II subunit alpha | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
7ZY2
 
 | | Crystal structure of compound 7 bound to CK2alpha | | Descriptor: | 5-bromanyl-1~{H}-indole, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, Fusco, C, Atkinson, E, Rossmann, M, Francis, N, Iegre, J, Hyvonen, M, Spring, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
6C8F
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of cesium | | Descriptor: | CESIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2MV3
 
 | | The N-domain of the AAA metalloproteinase Yme1 from Saccharomyces cerevisiae | | Descriptor: | Mitochondrial inner membrane i-AAA protease supercomplex subunit YME1 | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-22 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
5U5W
 
 | | CcP gateless cavity | | Descriptor: | 3-methylquinolin-4-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3MA9
 
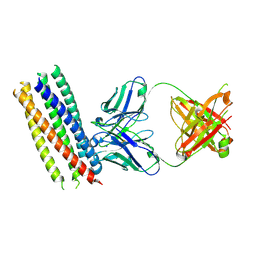 | | Crystal structure of gp41 derived protein complexed with fab 8066 | | Descriptor: | Fab8066 FAB ANTIBODY FRAGMENT, Heavy Chain, Light Chain, ... | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
6BZW
 
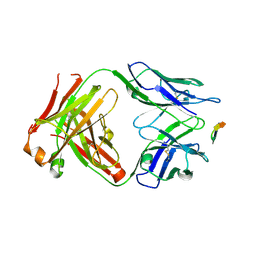 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2MTZ
 
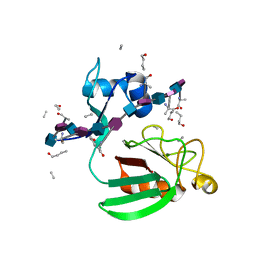 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
1KJ1
 
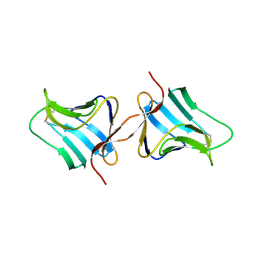 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | alpha-D-mannopyranose, lectin I, lectin II | | Authors: | Ramachandraiah, G, Chandra, N.R, Surolia, A, Vijayan, M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6BWO
 
 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, apo form | | Descriptor: | Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6BWV
 
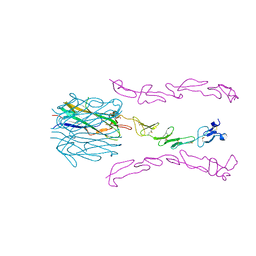 | | Crystal Structure of the 4-1BB/4-1BBL Complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Oganesyan, V, Gilbreth, R.N, Baca, M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human 4-1BB/4-1BBL complex.
J. Biol. Chem., 293, 2018
|
|
6BXO
 
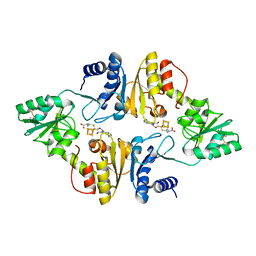 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
