6GM8
 
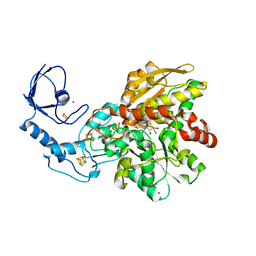 | | [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant E282Q | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | 著者 | Duan, J, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | 登録日 | 2018-05-24 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
8HMD
 
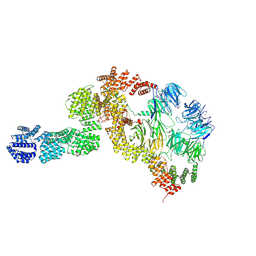 | | base module state 2 of Tetrahymena IFT-A | | 分子名称: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, Tetratricopeptide repeat protein, ... | | 著者 | Ma, Y, Wu, J, Lei, M. | | 登録日 | 2022-12-03 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
6RJU
 
 | |
8HME
 
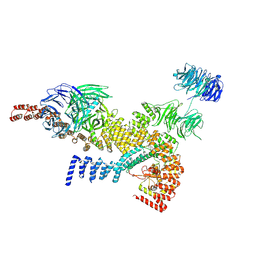 | | head module state 1 of Tetrahymena IFT-A | | 分子名称: | Intraflagellar transport protein 122 homolog, Intraflagellar transporter, WD40 repeat protein, ... | | 著者 | Ma, Y, Wu, J, Lei, M. | | 登録日 | 2022-12-03 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
8HMC
 
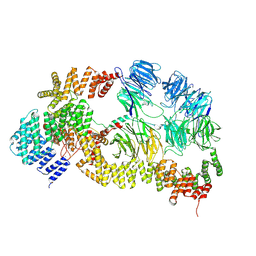 | | base module state 1 of Tetrahymena IFT-A | | 分子名称: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, Tetratricopeptide repeat protein, ... | | 著者 | Ma, Y, Wu, J, Lei, M. | | 登録日 | 2022-12-03 | | 公開日 | 2023-06-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insight into the intraflagellar transport complex IFT-A and its assembly in the anterograde IFT train.
Nat Commun, 14, 2023
|
|
8H1L
 
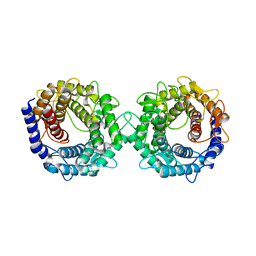 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | 分子名称: | N-acylglucosamine 2-epimerase, sorbitol | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6RGO
 
 | |
6GO5
 
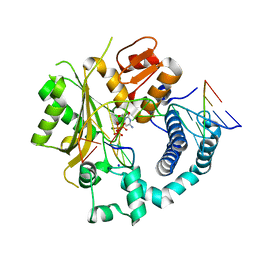 | | TdT chimera (Loop1 of pol mu) - Ternary complex with 1-nt gapped DNA substrate | | 分子名称: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(*AP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*GP*GP*CP*AP*AP*AP*CP*A)-3'), ... | | 著者 | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | 登録日 | 2018-06-01 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6RH3
 
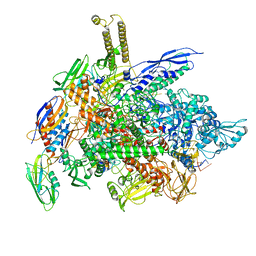 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | 登録日 | 2019-04-18 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
5G27
 
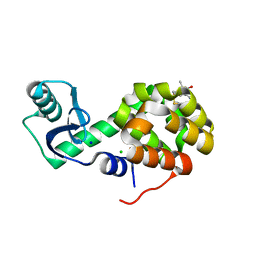 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at Room Temperature | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ENDOLYSIN, ... | | 著者 | Gohlke, U, Consentius, P, Loll, B, Mueller, R, Kaupp, M, Heinemann, U, Risse, T. | | 登録日 | 2016-04-07 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-Ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
8H1K
 
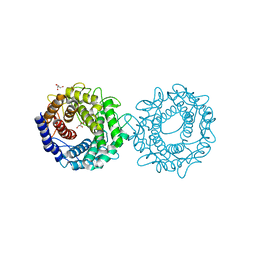 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3MSL
 
 | |
6RMJ
 
 | | Crystal structure of human NGR-TNF | | 分子名称: | Tumor necrosis factor | | 著者 | Degano, M, Garau, G. | | 登録日 | 2019-05-07 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mechanism of Action of the Tumor Vessel Targeting Agent NGR-hTNF: Role of Both NGR Peptide and hTNF in Cell Binding and Signaling.
Int J Mol Sci, 20, 2019
|
|
2QGM
 
 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | 分子名称: | Succinoglycan biosynthesis protein | | 著者 | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-06-29 | | 公開日 | 2007-07-24 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
5G5R
 
 | |
5JKK
 
 | |
6K0G
 
 | | Crystal Structure of UDP-glucose 4-epimerase from Bifidobacterium longum in complex with NAD+ and UDP | | 分子名称: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Nam, Y.-W, Nishimoto, M, Arakawa, T, Kitaoka, M, Fushinobu, S. | | 登録日 | 2019-05-06 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for broad substrate specificity of UDP-glucose 4-epimerase in the human milk oligosaccharide catabolic pathway of Bifidobacterium longum.
Sci Rep, 9, 2019
|
|
5MGI
 
 | | Crystal structure of KPC-2 carbapenemase in complex with a phenyl boronic inhibitor. | | 分子名称: | (~{E})-3-[2-(dihydroxyboranyl)phenyl]prop-2-enoic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | 著者 | Vicario, M, Celenza, G, Bellio, P, Perilli, M.G, Tondi, D, Cendron, L. | | 登録日 | 2016-11-21 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Phenylboronic Acid Derivatives as Validated Leads Active in Clinical Strains Overexpressing KPC-2: A Step against Bacterial Resistance.
Chemmedchem, 13, 2018
|
|
5MKF
 
 | | cryoEM Structure of Polycystin-2 in complex with calcium and lipids | | 分子名称: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wilkes, M, Madej, M.G, Ziegler, C. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7CIM
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
2QJG
 
 | | M. jannaschii ADH synthase complexed with F1,6P | | 分子名称: | 1,6-DI-O-PHOSPHONO-D-ALLITOL, Putative aldolase MJ0400 | | 著者 | Ealick, S.E, Morar, M. | | 登録日 | 2007-07-07 | | 公開日 | 2007-10-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid synthase, a catalyst in the archaeal pathway for the biosynthesis of aromatic amino acids.
Biochemistry, 46, 2007
|
|
6H7G
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | 分子名称: | Phosphoribulokinase, chloroplastic, SULFATE ION | | 著者 | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | 登録日 | 2018-07-31 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2QKS
 
 | |
8H1M
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | 分子名称: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | 著者 | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2022-10-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
