7CD1
 
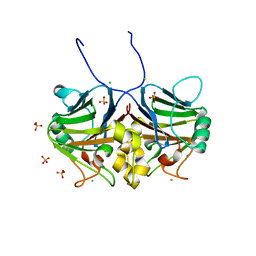 | | Crystal structure of inhibitory Smad, Smad7 | | 分子名称: | CHLORIDE ION, Mothers against decapentaplegic homolog 7, SULFATE ION | | 著者 | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for inhibitory effects of Smad7 on TGF-beta family signaling.
J.Struct.Biol., 212, 2020
|
|
6QQL
 
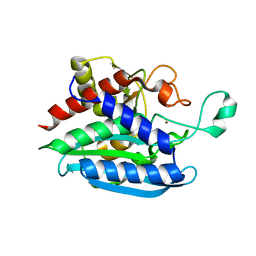 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | 分子名称: | Glutamine cyclotransferase, ZINC ION | | 著者 | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | 登録日 | 2019-02-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
3LG0
 
 | |
5VOT
 
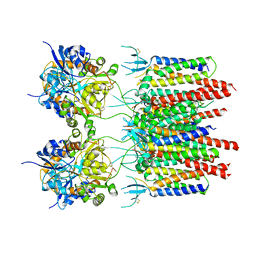 | | Structure of AMPA receptor-TARP complex | | 分子名称: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | 著者 | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | 登録日 | 2017-05-03 | | 公開日 | 2017-07-12 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5VPG
 
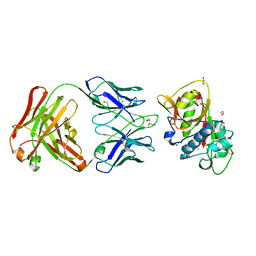 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | 登録日 | 2017-05-05 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
5VUG
 
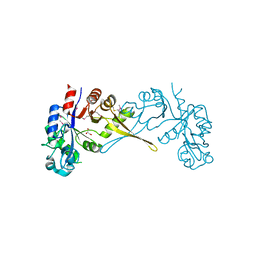 | | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis | | 分子名称: | CALCIUM ION, GLYCEROL, Uncharacterized protein Rv2277c | | 著者 | Kim, Y, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-14 | | 最終更新日 | 2017-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis
To Be Published
|
|
7NHZ
 
 | |
6KXE
 
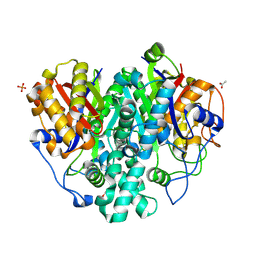 | | The ishigamide ketosynthase/chain length factor | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | 著者 | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | 登録日 | 2019-09-10 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
5VWK
 
 | |
4OBQ
 
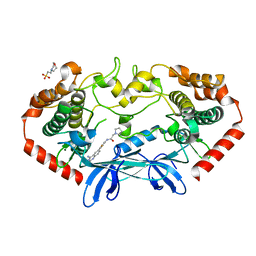 | | MAP4K4 in complex with inhibitor (compound 31), N-[3-(4-AMINOQUINAZOLIN-6-YL)-5-FLUOROPHENYL]-2-(PYRROLIDIN-1-YL)ACETAMIDE | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 4, ... | | 著者 | Harris, S.F, Wu, P, Coons, M. | | 登録日 | 2014-01-07 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery of Selective 4-Amino-pyridopyrimidine Inhibitors of MAP4K4 Using Fragment-Based Lead Identification and Optimization.
J.Med.Chem., 57, 2014
|
|
7CTN
 
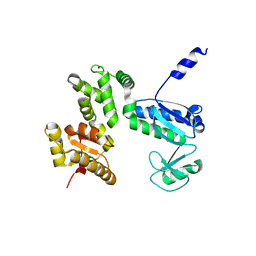 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | 分子名称: | Flagellar biosynthesis protein FlhA | | 著者 | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | 登録日 | 2020-08-19 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | 分子名称: | Heat shock protein 104 | | 著者 | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
5VPA
 
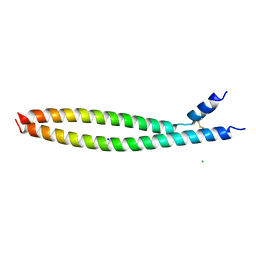 | | Transcription factor FosB/JunD bZIP domain | | 分子名称: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | 著者 | Yin, Z, Machius, M, Rudenko, G. | | 登録日 | 2017-05-04 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPF
 
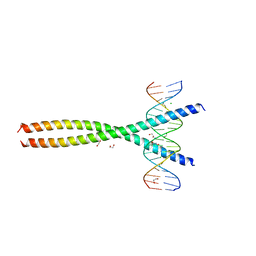 | | Transcription factor FosB/JunD bZIP domain in complex with cognate DNA, type-II crystal | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*GP*TP*CP*GP*GP*TP*GP*AP*CP*TP*CP*AP*CP*CP*GP*AP*CP*G)-3'), ... | | 著者 | Yin, Z, Rudenko, G, Machius, M. | | 登録日 | 2017-05-04 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.694 Å) | | 主引用文献 | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VWC
 
 | |
3L84
 
 | | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | 分子名称: | ACETATE ION, GLYCEROL, Transketolase | | 著者 | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-12-29 | | 公開日 | 2010-02-09 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | High resolution crystal structure of transketolase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
6R2X
 
 | | NMR structure of Chromogranin A (F39-D63) | | 分子名称: | Chromogranin-A | | 著者 | Nardelli, F, Quilici, G, Ghitti, M, Curnis, F, Gori, A, Berardi, A, Corti, A, Musco, G. | | 登録日 | 2019-03-19 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A stapled chromogranin A-derived peptide is a potent dual ligand for integrins alpha v beta 6 and alpha v beta 8.
Chem.Commun.(Camb.), 55, 2019
|
|
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | 著者 | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | 登録日 | 2020-06-08 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.341 Å) | | 主引用文献 | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
8P8P
 
 | | Crystal structure of human Histidine Triad Nucleotide-Binding Protein 1 in complex with 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl Ethenoadenosine | | 分子名称: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[2-(1~{H}-indol-3-yl)ethyl]carbamate | | 著者 | Dolot, R.M, Dillenburg, M, Wagner, C.R. | | 登録日 | 2023-06-02 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Novel inhibitors for hHINT1 protein
To Be Published
|
|
8PA9
 
 | |
3LE4
 
 | |
8PAI
 
 | |
6KXF
 
 | | The ishigamide ketosynthase/chain length factor | | 分子名称: | ACP, Ketosynthase, [(3~{R})-2,2-dimethyl-4-[[3-[2-[[(~{E})-oct-2-enoyl]amino]ethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] dihydrogen phosphate | | 著者 | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | 登録日 | 2019-09-10 | | 公開日 | 2020-05-06 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
7COK
 
 | | Crystal structure of ligand-free form of 5-ketofructose reductase of Gluconobacter sp. strain CHM43 | | 分子名称: | 5-ketofructose reductase | | 著者 | Noda, S, Hodoya, Y, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | 登録日 | 2020-08-04 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
7COL
 
 | | Crystal structure of 5-ketofructose reductase complexed with NADPH | | 分子名称: | 5-ketofructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Hodoya, Y, Noda, S, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | 登録日 | 2020-08-04 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
