6HWE
 
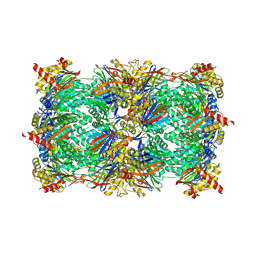 | | Yeast 20S proteasome beta2-G45A mutant in complex with carfilzomib | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-[[(3~{R},4~{S})-2,6-dimethyl-2,3-bis(oxidanyl)heptan-4-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]-4-methyl-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)-4-phenyl-butanoyl]amino]pentanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2018-10-11 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4XVU
 
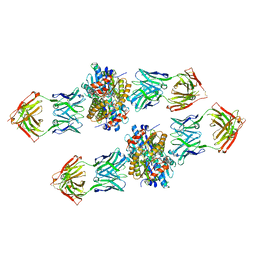 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | 著者 | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | 登録日 | 2015-01-28 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
8YA8
 
 | |
6I0N
 
 | |
4Y05
 
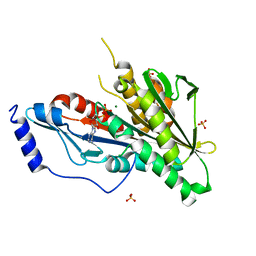 | | KIF2C short Loop2 construct | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION, ... | | 著者 | Wang, W, Knossow, M, Gigant, B. | | 登録日 | 2015-02-05 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | New Insights into the Coupling between Microtubule Depolymerization and ATP Hydrolysis by Kinesin-13 Protein Kif2C.
J.Biol.Chem., 290, 2015
|
|
7Z3B
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | 分子名称: | ACETATE ION, AcoP, COPPER (I) ION, ... | | 著者 | Leone, P, Sciara, G, Ilbert, M. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3F
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | 分子名称: | ACETATE ION, AcoP, CHLORIDE ION, ... | | 著者 | Leone, P, Sciara, G, Ilbert, M. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4Y0T
 
 | | Crystal structure of apo form of OXA-58, a Carbapenem hydrolyzing Class D beta-lactamase from Acinetobacter baumanii (P21, 4mol/ASU) | | 分子名称: | Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
7Z3G
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | 分子名称: | AcoP, COPPER (I) ION, GLYCEROL | | 著者 | Leone, P, Sciara, G, Ilbert, M. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | 分子名称: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | 登録日 | 1997-11-21 | | 公開日 | 1998-02-25 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
7Z3I
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | 分子名称: | ACETATE ION, AcoP, COPPER (II) ION, ... | | 著者 | Leone, P, Sciara, G, Ilbert, M. | | 登録日 | 2022-03-02 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
4Y1P
 
 | | Crystal structure of 3-isopropylmalate dehydrogenase (Saci_0600) from Sulfolobus acidocaldarius complex with 3-isopropylmalate and Mg2+ | | 分子名称: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | 著者 | Takahashi, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2015-02-08 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Characterization of two beta-decarboxylating dehydrogenases from Sulfolobus acidocaldarius
Extremophiles, 20, 2016
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4XA5
 
 | | Crystal structure of the pre-catalytic ternary complex of DNA polymerase lambda with a templating A and an incoming 8-oxo-dGTP | | 分子名称: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ACETATE ION, DNA polymerase lambda, ... | | 著者 | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | 登録日 | 2014-12-12 | | 公開日 | 2016-02-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda.
Nucleic Acids Res., 43, 2015
|
|
8XBX
 
 | | The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | 著者 | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | 登録日 | 1999-02-15 | | 公開日 | 1999-02-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4XAC
 
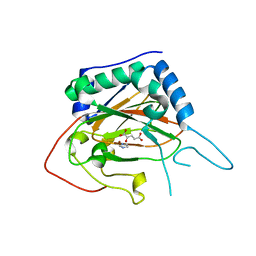 | | Crystal Structure of EvdO2 from Micromonospora carbonacea var. aurantiaca complexed with 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, EvdO2, IMIDAZOLE, ... | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XCA
 
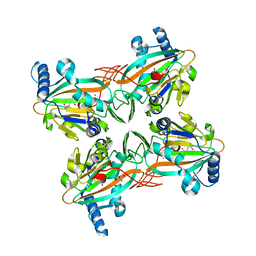 | | Crystal Structure of HygX from Streptomyces hygroscopicus with nickel and 2-oxoglutarate bound | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-OXOGLUTARIC ACID, CESIUM ION, ... | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1AOE
 
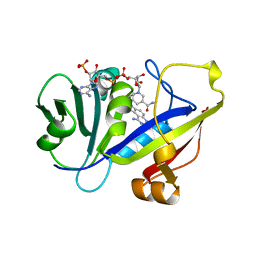 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | 分子名称: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Whitlow, M, Howard, A.J, Stewart, D. | | 登録日 | 1997-07-02 | | 公開日 | 1998-01-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
4XHS
 
 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | 分子名称: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | 著者 | Jin, T, Huang, M, Jiang, J, Xiao, T. | | 登録日 | 2015-01-06 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
6HTD
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 4 | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2018-10-03 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1B6X
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE GUANINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 4 STRUCTURES | | 分子名称: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3' | | 著者 | Cullinan, D, Johnson, F, Grollman, A.P, Eisenberg, M, De Los Santos, C. | | 登録日 | 1999-01-19 | | 公開日 | 1999-01-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a DNA duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyguanosine.
Biochemistry, 36, 1997
|
|
