8F67
 
 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | 分子名称: | Pbp5, SULFATE ION | | 著者 | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | 登録日 | 2022-11-16 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3T
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
6R2X
 
 | | NMR structure of Chromogranin A (F39-D63) | | 分子名称: | Chromogranin-A | | 著者 | Nardelli, F, Quilici, G, Ghitti, M, Curnis, F, Gori, A, Berardi, A, Corti, A, Musco, G. | | 登録日 | 2019-03-19 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A stapled chromogranin A-derived peptide is a potent dual ligand for integrins alpha v beta 6 and alpha v beta 8.
Chem.Commun.(Camb.), 55, 2019
|
|
8F3I
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Z
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S422A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3U
 
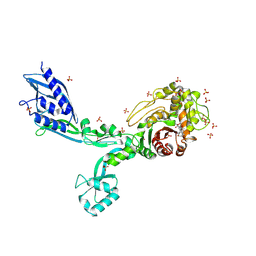 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3N
 
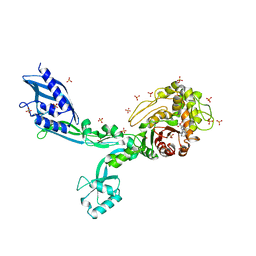 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3F
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Hunashal, Y, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
4MY6
 
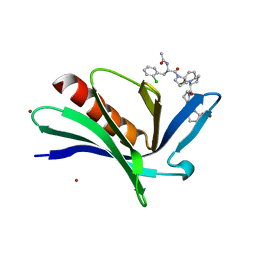 | | EnaH-EVH1 in complex with peptidomimetic low-molecular weight inhibitor Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-OH | | 分子名称: | (3aR,5aS,8S,10aS)-1-[(3S,6R,8aS)-1'-[(2S)-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8a-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-10-oxidanylidene-2,3,3a,5a,8,10a-hexahydrodipyrrolo[3,2-b:3',1'-f]azepine-8-carboxylic acid, BROMIDE ION, Protein enabled homolog | | 著者 | Barone, M, Roske, Y, Kuehne, R. | | 登録日 | 2013-09-27 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A modular toolkit to inhibit proline-rich motif-mediated protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8F3R
 
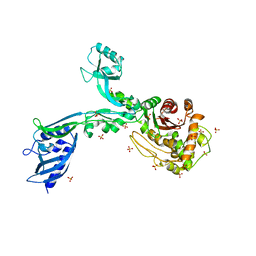 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
3CZD
 
 | | Crystal structure of human glutaminase in complex with L-glutamate | | 分子名称: | GLUTAMIC ACID, GLYCEROL, Glutaminase kidney isoform, ... | | 著者 | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-04-29 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8F3P
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) R464A variant penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3J
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3M
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485A variant with S466 insertion apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3G
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant in the penicillin bound form from Enterococcus faecium | | 分子名称: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Q
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Y460A variant apo form from Enterococcus faecium | | 分子名称: | Penicillin binding protein 5, SULFATE ION | | 著者 | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Page, R, Peti, W. | | 登録日 | 2022-11-10 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | 著者 | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | 登録日 | 2023-07-24 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8CSE
 
 | | WbbB in complex with alpha-Rha-(1-3)-beta-GlcNAc acceptor | | 分子名称: | CYTIDINE-5'-MONOPHOSPHATE, N-(8-hydroxyoctyl)-4-methoxybenzamide, N-acetyl glucosaminyl transferase, ... | | 著者 | Forrester, T.J.B, Kimber, M.S. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSF
 
 | |
8CSD
 
 | | WbbB D232C Kdo adduct | | 分子名称: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Forrester, T.J.B, Kimber, M.S. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSB
 
 | | WbbB D232N in complex with CMP-beta-Kdo | | 分子名称: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, CYTIDINE-5'-MONOPHOSPHATE, N-acetyl glucosaminyl transferase, ... | | 著者 | Forrester, T.J.B, Kimber, M.S. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
8CSC
 
 | | WbbB D232N-Kdo adduct | | 分子名称: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, CHLORIDE ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Forrester, T.J.B, Kimber, M.S. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The retaining beta-Kdo glycosyltransferase WbbB uses a double-displacement mechanism with an intermediate adduct rearrangement step.
Nat Commun, 13, 2022
|
|
1A03
 
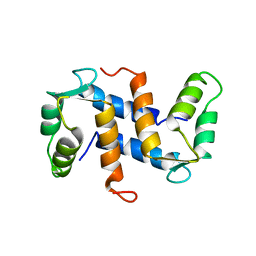 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | 分子名称: | CALCYCLIN (RABBIT, CA2+) | | 著者 | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | 登録日 | 1997-12-08 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
8T88
 
 | |
6R64
 
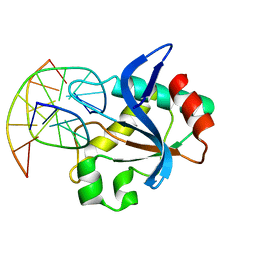 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | 分子名称: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | 著者 | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | 登録日 | 2019-03-26 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
