3ZS5
 
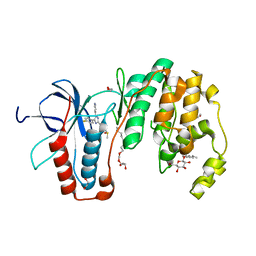 | | Structural basis for kinase selectivity of three clinical p38alpha inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MITOGEN-ACTIVATED PROTEIN KINASE 14, ... | | 著者 | Azevedo, R, van Zeeland, M, Raaijmakers, H.C.A, Kazemier, B, Oubrie, A. | | 登録日 | 2011-06-23 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
2L4N
 
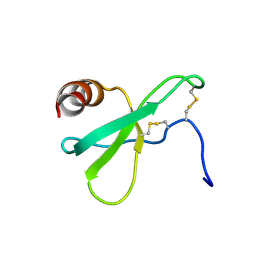 | |
3X2L
 
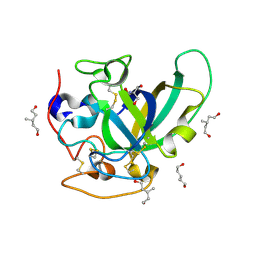 | | X-ray structure of PcCel45A apo form at 95K. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | 著者 | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (0.83 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3ZJE
 
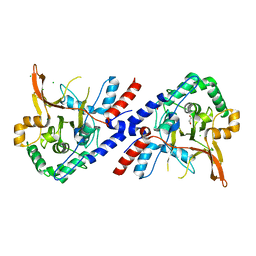 | | A20 OTU domain in reversibly oxidised (SOH) state | | 分子名称: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | 著者 | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | 登録日 | 2013-01-17 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
4F60
 
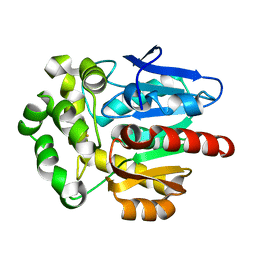 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | 分子名称: | FLUORIDE ION, Haloalkane dehalogenase | | 著者 | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | 登録日 | 2012-05-14 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3ZK8
 
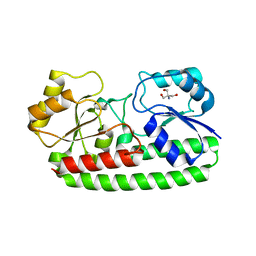 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA E205Q IN THE METAL-FREE, OPEN STATE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | 著者 | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | 登録日 | 2013-01-22 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
2LA8
 
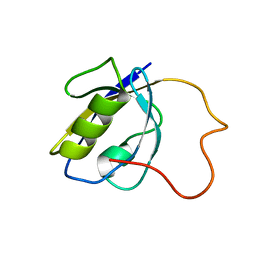 | |
2LGE
 
 | |
4XL5
 
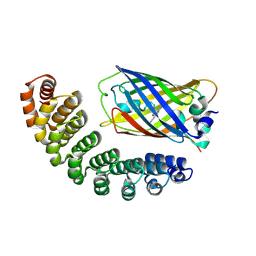 | | X-ray structure of bGFP-A / EGFP complex | | 分子名称: | Green fluorescent protein, bGFP-A | | 著者 | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | 登録日 | 2015-01-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
2LM3
 
 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | 分子名称: | Tripartite motif-containing protein 5 | | 著者 | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | 登録日 | 2011-11-21 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LLL
 
 | | Solution NMR structure of C-terminal globular domain of human Lamin-B2, Northeast Structural Genomics Consortium target HR8546A | | 分子名称: | Lamin-B2 | | 著者 | Lemak, A, Yee, A, Houliston, S, Garcia, M, Xu, C, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | 登録日 | 2011-11-11 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of c-terminal globular domain of human Lamin-B2
To be Published
|
|
3ZDG
 
 | | Crystal Structure of Ls-AChBP complexed with carbamoylcholine analogue 3-(dimethylamino)butyl dimethylcarbamate (DMABC) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(dimethylamino)butyl dimethylcarbamate, ACETYLCHOLINE BINDING PROTEIN, ... | | 著者 | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | 登録日 | 2012-11-26 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
1MO0
 
 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | 分子名称: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | 著者 | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2002-09-06 | | 公開日 | 2002-09-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
2LQH
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | 分子名称: | CREB-binding protein, Forkhead box O3 | | 著者 | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | 登録日 | 2012-03-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZFS
 
 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | 著者 | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | 登録日 | 2012-12-12 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
2L29
 
 | | Complex structure of E4 mutant human IGF2R domain 11 bound to IGF-II | | 分子名称: | Insulin-like growth factor 2 receptor variant, Insulin-like growth factor II | | 著者 | Williams, C, Hoppe, H, Rezgui, D, Strickland, M, Frago, S, Ellis, R.Z, Wattana-Amorn, P, Prince, S.N, Zaccheo, O.J, Forbes, B, Jones, E.Y, Crump, M.P, Hassan, A.B. | | 登録日 | 2010-08-13 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
3ZHC
 
 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | 分子名称: | CHLORIDE ION, FORMIC ACID, PHYTASE | | 著者 | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | 登録日 | 2012-12-20 | | 公開日 | 2013-08-28 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
3ZVG
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | 分子名称: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | 著者 | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2011-07-24 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | 分子名称: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | 著者 | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | 登録日 | 2022-06-30 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
2L4E
 
 | |
3X0E
 
 | |
2LRV
 
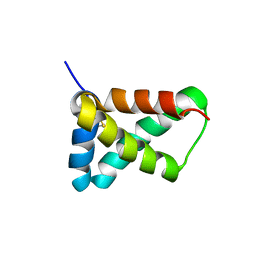 | |
3X0G
 
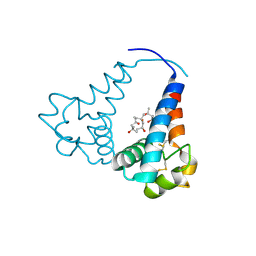 | |
2LK0
 
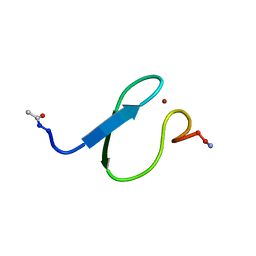 | |
3X2N
 
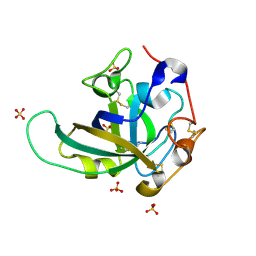 | | Proton relay pathway in inverting cellulase | | 分子名称: | Endoglucanase V-like protein, SULFATE ION | | 著者 | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | 登録日 | 2014-12-22 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
