3FL1
 
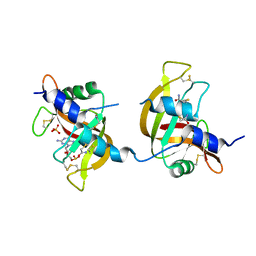 | | X-ray structure of the non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | 分子名称: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic, SULFATE ION, ... | | 著者 | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | 登録日 | 2008-12-18 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
3RIS
 
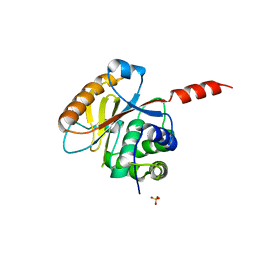 | | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme | | 分子名称: | GLYCEROL, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | 著者 | Das, C, Permaul, M, Maiti, T.K. | | 登録日 | 2011-04-14 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | Crystal structure of the catalytic domain of UCHL5, a proteasome-associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme.
Febs J., 278, 2011
|
|
6G88
 
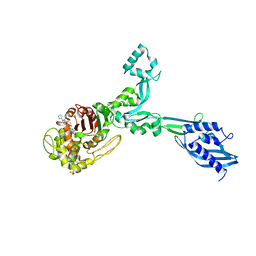 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | 著者 | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | 登録日 | 2018-04-08 | | 公開日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
4NXW
 
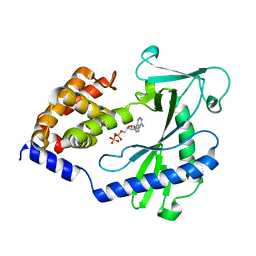 | |
3ROE
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine | | 分子名称: | Apolipoprotein A-I-binding protein, THYMIDINE | | 著者 | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | 登録日 | 2011-04-25 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3F9R
 
 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | 分子名称: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | 著者 | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | 登録日 | 2008-11-14 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
6TO4
 
 | |
4L5K
 
 | | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine | | 分子名称: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | 著者 | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | 登録日 | 2013-06-11 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine
to be published
|
|
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | 分子名称: | chitinase | | 著者 | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | 登録日 | 2005-07-13 | | 公開日 | 2006-07-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
3RQX
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P4-Di(adenosine-5') tetraphosphate | | 分子名称: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-04-28 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRB
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, POTASSIUM ION, ... | | 著者 | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | 登録日 | 2011-04-29 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3FBG
 
 | | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus | | 分子名称: | MAGNESIUM ION, putative arginate lyase | | 著者 | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-11-19 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus
To be Published
|
|
4NPV
 
 | | Crystal structure of human PDE1B bound to inhibitor 7A (6,7,8-trimethoxy-N-(pentan-3-yl)quinazolin-4-amine) | | 分子名称: | 6,7,8-trimethoxy-N-(pentan-3-yl)quinazolin-4-amine, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | 著者 | Pandit, J, Evdomikov, A, Mansour, M, Simons, S. | | 登録日 | 2013-11-22 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Small-molecule phosphodiesterase probes: discovery of potent and selective CNS-penetrable quinazoline inhibitors of PDE1
MEDCHEMCOMM, 2014
|
|
3FNR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | 分子名称: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-12-26 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
6TOE
 
 | |
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | 分子名称: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Yu, A, Xie, Y, Li, M. | | 登録日 | 2019-07-05 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
6E0W
 
 | | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid | | 分子名称: | 1,2-ETHANEDIOL, 1,5-anhydro-4,6-O-[(1R)-1-carboxyethylidene]-D-galactitol, Bacteriophage Phi92 gp150, ... | | 著者 | Plattner, M, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Leiman, P.G, Schwarzer, D. | | 登録日 | 2018-07-07 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
6TPE
 
 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | 分子名称: | 2-[4-(3-methyl-6-oxidanylidene-1,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | 著者 | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Jestel, A, Lammens, A, Ritzen, A. | | 登録日 | 2019-12-13 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-07-22 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
3FOL
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFERI | | 分子名称: | 8 residue synthetic peptide, Beta-2-microglobulin, MHC | | 著者 | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | 登録日 | 2008-12-30 | | 公開日 | 2010-01-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
Int.Immunol., 22, 2010
|
|
4L7P
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor ME0395 | | 分子名称: | (2E)-N-[(2S)-1-hydroxy-3-phenylpropan-2-yl]-3-(4-oxo-1,4-dihydroquinazolin-2-yl)prop-2-enamide, Poly [ADP-ribose] polymerase 3 | | 著者 | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | 登録日 | 2013-06-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
6TQW
 
 | |
1KH1
 
 | |
4NQU
 
 | | anti-parallel Fc-knob (T366W) homodimer | | 分子名称: | Ig gamma-1 chain C region, SULFATE ION | | 著者 | Eigenbrot, C, Ultsch, M. | | 登録日 | 2013-11-25 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Antiparallel Conformation of Knob and Hole Aglycosylated Half-Antibody Homodimers Is Mediated by a CH2-CH3 Hydrophobic Interaction.
J.Mol.Biol., 426, 2014
|
|
4L90
 
 | | Crystal structure of Human Hsp90 with RL3 | | 分子名称: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal structure of Human Hsp90 with RL3
to be published
|
|
4KUA
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 | | 分子名称: | 1,2-ETHANEDIOL, GNAT superfamily acetyltransferase PA4794, SULFATE ION | | 著者 | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-05-21 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa.
J.Biol.Chem., 288, 2013
|
|
