7Z6M
 
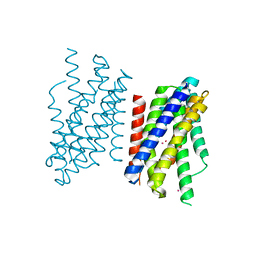 | | Crystal structure of Zn2+-transporter BbZIP in a cadmium bound state | | 分子名称: | CADMIUM ION, Putative membrane protein | | 著者 | Wiuf, A, Steffen, J.H, Becares, E.R, Groenberg, C, Mahato, D.R, Rasmussen, S.G.F, Andersson, M, Croll, T, Gotfryd, K, Gourdon, P. | | 登録日 | 2022-03-13 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | The two-domain elevator-type mechanism of zinc-transporting ZIP proteins.
Sci Adv, 8, 2022
|
|
1IN3
 
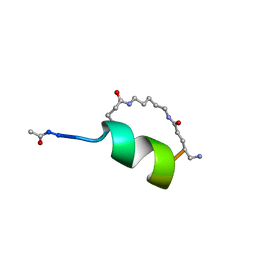 | | Peptide Antagonist of IGFBP1, (i,i+8) Covalently Restrained Analog | | 分子名称: | IGFBP-1 antagonist, PENTANE | | 著者 | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | 登録日 | 2001-05-11 | | 公開日 | 2001-05-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
6EOU
 
 | |
4L6Z
 
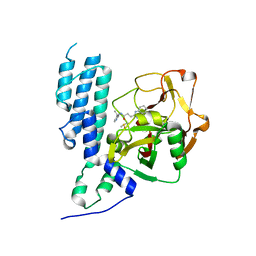 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor STO1168 | | 分子名称: | 3-(4-oxo-3,4-dihydroquinazolin-2-yl)-N-[(1S)-1-(pyridin-4-yl)ethyl]propanamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 3 | | 著者 | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | 登録日 | 2013-06-13 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
1IT6
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | 分子名称: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | 著者 | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | 登録日 | 2002-01-09 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
4LDU
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | 分子名称: | Auxin response factor 5, CHLORIDE ION | | 著者 | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | 登録日 | 2013-06-25 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4L7O
 
 | | Human artd3 (parp3) - catalytic domain in complex with inhibitor STO1542 | | 分子名称: | DIMETHYL SULFOXIDE, N-{(1S)-1-[4-(1H-imidazol-1-yl)phenyl]ethyl}-3-(4-oxo-3,4-dihydroquinazolin-2-yl)propanamide, Poly [ADP-ribose] polymerase 3 | | 著者 | Karlberg, T, Thorsell, A.G, Lindgren, A.E.G, Ekblad, T, Spjut, S, Andersson, C.D, Weigelt, J, Linusson, A, Elofsson, M, Schuler, H. | | 登録日 | 2013-06-14 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chemical Probes to Study ADP-Ribosylation: Synthesis and Biochemical Evaluation of Inhibitors of the Human ADP-Ribosyltransferase ARTD3/PARP3.
J.Med.Chem., 56, 2013
|
|
7ZF1
 
 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | 分子名称: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | 著者 | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | 登録日 | 2022-03-31 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.14 Å) | | 主引用文献 | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
6ISV
 
 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | 分子名称: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | 登録日 | 2018-11-19 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
4ZUK
 
 | | Structure ALDH7A1 complexed with NAD+ | | 分子名称: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | 著者 | Luo, M, Tanner, J.J. | | 登録日 | 2015-05-16 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
1IXK
 
 | | Crystal Structure Analysis of Methyltransferase Homolog Protein from Pyrococcus Horikoshii | | 分子名称: | Methyltransferase | | 著者 | Ishikawa, I, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | 登録日 | 2002-06-25 | | 公開日 | 2003-09-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
1IY0
 
 | | Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus | | 分子名称: | ATP-dependent metalloprotease FtsH, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | 登録日 | 2002-07-10 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
6E6R
 
 | | 1.50 A resolution structure of the C-terminally truncated [2Fe-2S] ferredoxin (Bfd) R26E mutant from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin-associated ferredoxin, FE2/S2 (INORGANIC) CLUSTER | | 著者 | Lovell, S, Wijerathne, H, Battaile, K.P, Yao, H, Wang, Y, Rivera, M. | | 登録日 | 2018-07-25 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Bfd, a New Class of [2Fe-2S] Protein That Functions in Bacterial Iron Homeostasis, Requires a Structural Anion Binding Site.
Biochemistry, 57, 2018
|
|
6IQG
 
 | | X-ray crystal structure of Fc and peptide complex | | 分子名称: | 18-mer peptide G(HCS)DCAYHRGELVWCT(HCS)H(NH2), 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Adachi, M, Ito, Y. | | 登録日 | 2018-11-08 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.998 Å) | | 主引用文献 | Site-Specific Chemical Conjugation of Antibodies by Using Affinity Peptide for the Development of Therapeutic Antibody Format.
Bioconjug. Chem., 30, 2019
|
|
6IUH
 
 | |
5ADO
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - Light chain S35R mutant | | 分子名称: | DIETHYL PHOSPHONATE, FAB A.17 | | 著者 | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | 登録日 | 2015-08-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
1IOR
 
 | | STABILIZATION OF HEN EGG WHITE LYSOZYME BY A CAVITY-FILLING MUTATION | | 分子名称: | LYSOZYME C | | 著者 | Ohmura, T, Ueda, T, Ootsuka, K, Saito, M, Imoto, T. | | 登録日 | 2001-03-28 | | 公開日 | 2001-04-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Stabilization of hen egg white lysozyme by a cavity-filling mutation.
Protein Sci., 10, 2001
|
|
3LVG
 
 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | 分子名称: | Clathrin heavy chain 1, Clathrin light chain B | | 著者 | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | 登録日 | 2010-02-20 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (7.94 Å) | | 主引用文献 | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
5AFP
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with rhodopsin kinase peptide from Homo sapiens | | 分子名称: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, RHODOPSIN KINASE, ... | | 著者 | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | 登録日 | 2015-01-23 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
1J1V
 
 | | Crystal structure of DnaA domainIV complexed with DnaAbox DNA | | 分子名称: | 5'-D(*CP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*G)-3', Chromosomal replication initiator protein dnaA | | 著者 | Fujikawa, N, Kurumizaka, H, Nureki, O, Terada, T, Shirouzu, M, Katayama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-12-18 | | 公開日 | 2003-04-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of replication origin recognition by the DnaA protein
NUCLEIC ACIDS RES., 31, 2003
|
|
3LWK
 
 | | Crystal structure of human Beta-crystallin A4 (CRYBA4) | | 分子名称: | Beta-crystallin A4, GLYCEROL, PHOSPHATE ION | | 著者 | Chaikuad, A, Shafqat, N, Krojer, T, Yue, W.W, Cocking, R, Vollmar, M, Muniz, J.R.C, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2010-02-24 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of human Beta-crystallin A4 (CRYBA4)
To be Published
|
|
6ITB
 
 | |
5AHL
 
 | | Apo-form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | 分子名称: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, SODIUM ION | | 著者 | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | 登録日 | 2015-02-06 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5AHK
 
 | | Crystal structure of acetohydroxy acid synthase Pf5 from Pseudomonas protegens | | 分子名称: | ACETOLACTATE SYNTHASE II, LARGE SUBUNIT, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Dobritzsch, D, Loschonsky, S, Mueller, M, Schneider, G. | | 登録日 | 2015-02-06 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The Crystal Structure of the Acetohydroxy Acid Synthase Pf5 from Pseudomonas Protegens
To be Published
|
|
