3X0A
 
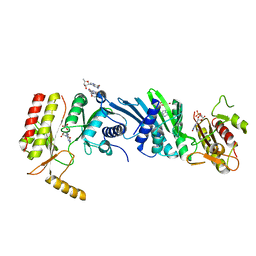 | | Crystal structure of PIP4KIIBETA F205L complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X25
 
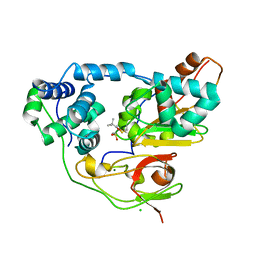 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | 登録日 | 2014-12-10 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3ZSC
 
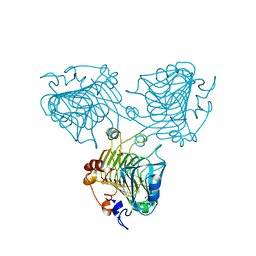 | | Catalytic function and substrate recognition of the pectate lyase from Thermotoga maritima | | 分子名称: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, GLYCEROL, PECTATE TRISACCHARIDE-LYASE, ... | | 著者 | McDonough, M.A, Thymark, M, Frisner, H, Hotchkiss, A, Sonksen, C, Bjornvad, M, Johansen, K.S, Larsen, S. | | 登録日 | 2011-06-24 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Catalytic Function and Substrate Recognition of the Pectate Lyase from Thermotoga Maritima
To be Published
|
|
1LJG
 
 | |
1LLC
 
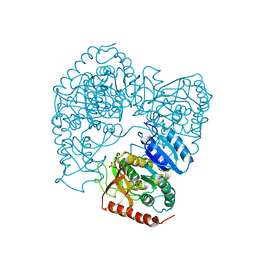 | | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS CASEI AT 3.0 ANGSTROMS RESOLUTION | | 分子名称: | 1,6-di-O-phosphono-alpha-D-fructofuranose, L-LACTATE DEHYDROGENASE, SULFATE ION | | 著者 | Buehner, M, Hecht, H.J, Hensel, R. | | 登録日 | 1988-11-21 | | 公開日 | 1989-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | STRUCTURE DETERMINATION OF THE ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS-CASEI AT 3A RESOLUTION.
Acta Crystallogr.,Sect.A, 40, 1984
|
|
2L3Z
 
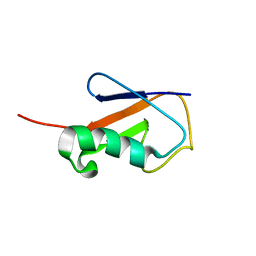 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | 分子名称: | Ubiquitin | | 著者 | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | 登録日 | 2010-09-27 | | 公開日 | 2011-02-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
3ZI5
 
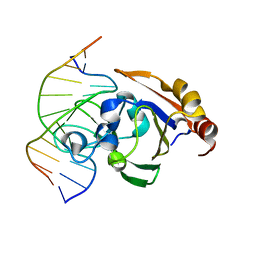 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | 分子名称: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | 著者 | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | 登録日 | 2013-01-03 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
3WZP
 
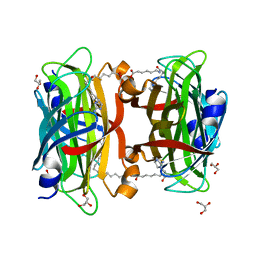 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | 分子名称: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X0C
 
 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0B
 
 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X20
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 25 min | | 分子名称: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | 著者 | Yamanaka, Y, Hashimoto, K, Noguchi, N, Yohda, M, Odaka, M. | | 登録日 | 2014-12-03 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4M3S
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with HEPES | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | 著者 | Majorek, K.A, Chruszcz, M, Xu, X, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-08-06 | | 公開日 | 2013-08-21 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments.
Protein Sci., 23, 2014
|
|
7CJR
 
 | | Crystal structure of a periplasmic sensor domain of histidine kinase VbrK | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidine kinase | | 著者 | Goh, B.C, Chua, Y.K, Qian, X, Savko, M, Lescar, J. | | 登録日 | 2020-07-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2020-10-14 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of the periplasmic sensor domain of histidine kinase VbrK suggests indirect sensing of beta-lactam antibiotics.
J.Struct.Biol., 212, 2020
|
|
2LKN
 
 | | Solution structure of the PPIase domain of human aryl-hydrocarbon receptor-interacting protein (AIP) | | 分子名称: | AH receptor-interacting protein | | 著者 | Linnert, M, Lin, Y, Manns, A, Haupt, K, Paschke, A, Fischer, G, Weiwad, M, Luecke, C. | | 登録日 | 2011-10-17 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The FKBP-Type Domain of the Human Aryl Hydrocarbon Receptor-Interacting Protein Reveals an Unusual Hsp90 Interaction.
Biochemistry, 52, 2013
|
|
1J0T
 
 | | The solution structure of molt-inhibiting hormone from the kuruma prawn | | 分子名称: | MOLT-INHIBITING HORMONE | | 著者 | Katayama, H, Nagata, K, Ohira, T, Yumoto, F, Tanokura, M, Nagasawa, H. | | 登録日 | 2002-11-22 | | 公開日 | 2002-12-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of molt-inhibiting hormone from the Kuruma prawn Marsupenaeus japonicus
J.Biol.Chem., 278, 2003
|
|
3WZQ
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | 分子名称: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
2LLU
 
 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | 分子名称: | Protein S100-A1 | | 著者 | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | 登録日 | 2011-11-17 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
2LLH
 
 | | NMR structure of Npm1_c70 | | 分子名称: | Nucleophosmin | | 著者 | Banci, L, Bertini, I, Brunori, M, Di Matteo, A, Federici, L, Gallo, A, Lo Sterzo, C, Mori, M. | | 登録日 | 2011-11-09 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter.
J.Biol.Chem., 287, 2012
|
|
3T1U
 
 | | Crystal Structure of the complex of Cyclophilin-A enzyme from Azotobacter vinelandii with sucAFPFpNA peptide | | 分子名称: | Peptidyl-prolyl cis-trans isomerase, succinyl-Ala-Phe-Pro-Phe-p-nitroanilide | | 著者 | Karpusas, M, Christoforides, E, Bethanis, K, Dimou, M, Katinakis, P. | | 登録日 | 2011-07-22 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of a bacterial cytoplasmic cyclophilin A in complex with a tetrapeptide.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3T41
 
 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | 分子名称: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | 著者 | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-25 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
2PJR
 
 | | HELICASE PRODUCT COMPLEX | | 分子名称: | DNA (5'-D(*AP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*T)-3'), ... | | 著者 | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | 登録日 | 1999-03-12 | | 公開日 | 1999-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.
Cell(Cambridge,Mass.), 97, 1999
|
|
2PZS
 
 | | Phi29 DNA polymerase complexed with primer-template DNA (post-translocation binary complex) | | 分子名称: | 5'-d(CTAACACGTAAGCAGTC)-3', 5'-d(GACTGCTTAC)-3', DNA polymerase | | 著者 | Berman, A.J, Kamtekar, S, Goodman, J.L, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | 登録日 | 2007-05-18 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases
Embo J., 26, 2007
|
|
3T6W
 
 | | Crystal Structure of Steccherinum ochraceum Laccase obtained by multi-crystals composite data collection technique (10% dose) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | Ferraroni, M, Briganti, F, Matera, I, Kolomytseva, M, Golovleva, L, Scozzafava, A, Chernykh, A.M. | | 登録日 | 2011-07-29 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Reaction intermediates and redox state changes in a blue laccase from Steccherinum ochraceum observed by crystallographic high/low X-ray dose experiments.
J.Inorg.Biochem., 111, 2012
|
|
1IVB
 
 | |
3SEP
 
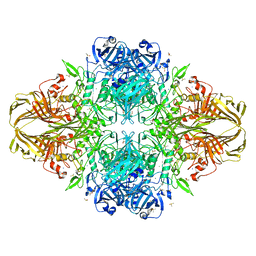 | | E. coli (lacZ) beta-galactosidase (S796A) | | 分子名称: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | 登録日 | 2011-06-10 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
