6SEC
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG | | Descriptor: | 2-nitrophenyl beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.768 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6EY2
 
 | | Crystal structure of XIAP-BIR3 in complex with a cIAP1-selective SM | | Descriptor: | (3~{S},6~{S},7~{S},9~{a}~{S})-~{N}-[(4-~{tert}-butylphenyl)methyl]-7-(hydroxymethyl)-6-[[(2~{S})-2-(methylamino)butanoyl]amino]-5-oxidanylidene-1,2,3,6,7,8,9,9~{a}-octahydropyrrolo[1,2-a]azepine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Cossu, F, Corti, A, Milani, M, Mastrangelo, E. | | Deposit date: | 2017-11-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design and molecular profiling of Smac-mimetics selective for cellular IAPs.
FEBS J., 285, 2018
|
|
5M67
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Bradyrhizobium elkanii in complex with adenine and 2'-deoxyadenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, ACETATE ION, ADENINE, ... | | Authors: | Manszewski, T, Jaskolski, M. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallographic and SAXS studies of S-adenosyl-l-homocysteine hydrolase from Bradyrhizobium elkanii.
IUCrJ, 4, 2017
|
|
2QO5
 
 | | Crystal structure of the cysteine 91 threonine mutant of zebrafish liver bile acid-binding protein complexed with cholic acid | | Descriptor: | CHOLIC ACID, Liver-basic fatty acid binding protein | | Authors: | Capaldi, S, Saccomani, G, Perduca, M, Monaco, H.L. | | Deposit date: | 2007-07-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Single Amino Acid Mutation in Zebrafish (Danio rerio) Liver Bile Acid-binding Protein Can Change the Stoichiometry of Ligand Binding.
J.Biol.Chem., 282, 2007
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
5F2T
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with palmitate - C 2 space group | | Descriptor: | MAGNESIUM ION, PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
6ZVV
 
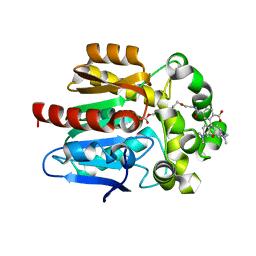 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVT
 
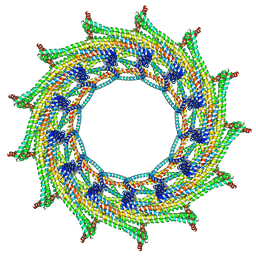 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
5MPP
 
 | | Structure of AaLS-wt | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
2QPW
 
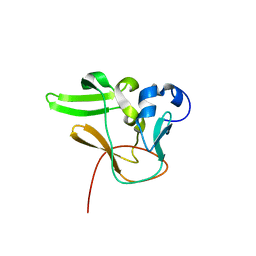 | | Methyltransferase domain of human PR domain-containing protein 2 | | Descriptor: | PR domain zinc finger protein 2 | | Authors: | Lunin, V.V, Wu, H, Dombrovski, L, Antoshenko, T, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
6ZW7
 
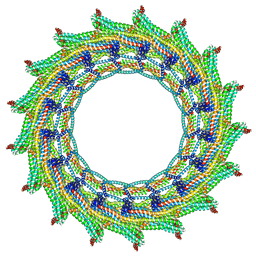 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6SAZ
 
 | | Cleaved human fetuin-b in complex with crayfish astacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Cuppari, A, Korschgen, H, Schmitz, C, Kuske, M, Yiallouros, I, Floehr, J, Jahnen-Dechent, W, Stocker, W. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases.
Sci Rep, 9, 2019
|
|
6F71
 
 | |
6ZVW
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVU
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6ZVX
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165H-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6SE8
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
2QC5
 
 | | Streptogramin B lyase structure | | Descriptor: | IODIDE ION, Streptogramin B lactonase | | Authors: | Lipka, M, Bochtler, M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of the Staphylococcus cohnii virginiamycin B lyase (Vgb).
Biochemistry, 47, 2008
|
|
6SEA
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | Descriptor: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SFE
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
1I35
 
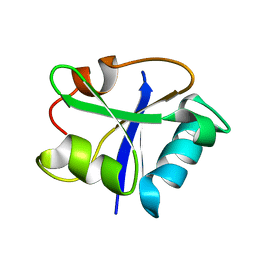 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF THE PROTEIN KINASE BYR2 FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PROTEIN KINASE BYR2 | | Authors: | Gronwald, W, Huber, F, Grunewald, P, Sporner, M, Wohlgemuth, S, Herrmann, C, Kalbitzer, H.R. | | Deposit date: | 2001-02-13 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe.
Structure, 9, 2001
|
|
6F4F
 
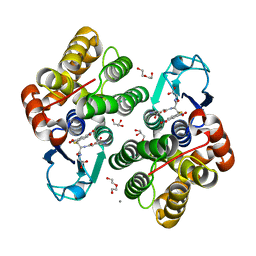 | | Crystal structure of glutathione transferase Omega 3S from Trametes versicolor in complex with glutathionyl-S-dinitrobenzene | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLUTATHIONE S-(2,4 DINITROBENZENE), ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
2LT2
 
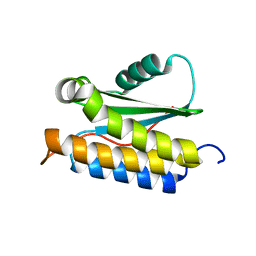 | |
