5JEP
 
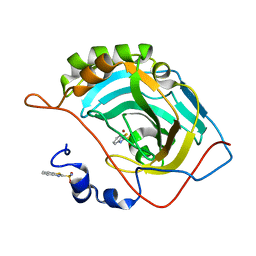 | | Human carbonic anhydrase II (T199S) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5IY0
 
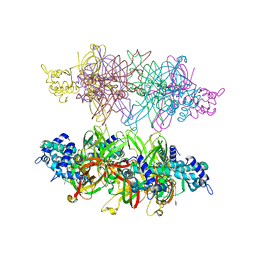 | | PfMCM N-terminal domain double hexamer | | Descriptor: | Cell division control protein 21, ZINC ION | | Authors: | Meagher, M, Enemark, E.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a double hexamer of the Pyrococcus furiosus minichromosome maintenance protein N-terminal domain.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5JG7
 
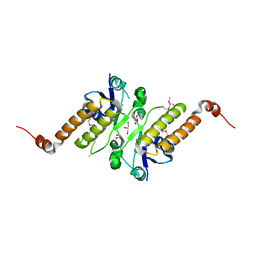 | | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2 | | Descriptor: | Fur regulated Salmonella iron transporter, GLYCEROL | | Authors: | Chang, C, Zhou, M, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-19 | | Release date: | 2016-04-27 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative periplasmic binding protein from Salmonella typhimurium LT2
To Be Published
|
|
5JBJ
 
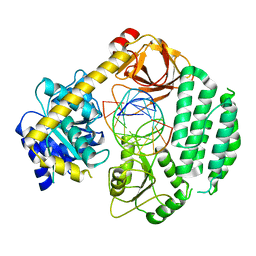 | |
5JBR
 
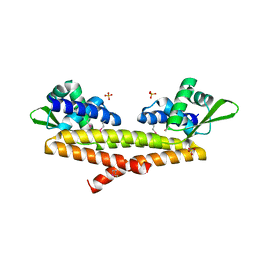 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | Descriptor: | SULFATE ION, Uncharacterized protein Bcav_2135 | | Authors: | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-04-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
5JBV
 
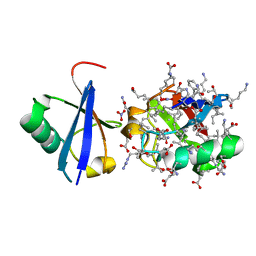 | | Lys27-linked triubiquitin | | Descriptor: | D-ubiquitin, NITRATE ION, Ubiquitin | | Authors: | Pan, M, Gao, S, Zheng, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structure of Lys27-linked triubiquitin
To Be Published
|
|
5JCV
 
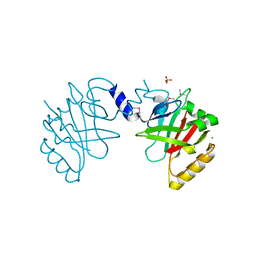 | | Sortase B from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2181 protein, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-04-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Sortase B from Listeria monocytogenes.
to be published
|
|
5J0F
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P4/5 | | Descriptor: | GLYCEROL, Superoxide dismutase [Cu-Zn],OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5JFC
 
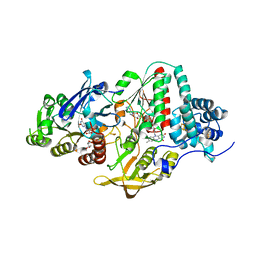 | | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, King, P.W, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
5J3U
 
 | | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Protein Kinase A | | Authors: | El Bakkouri, M, Walker, J.R, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP
To Be Published
|
|
5J4X
 
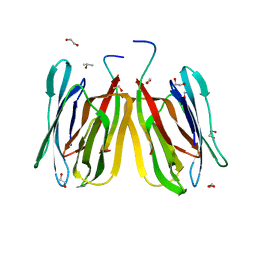 | | Structure of tetrameric jacalin complexed with Gal beta-(1,3) Gal-beta-OMe | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Surolia, A, Vijayan, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Distortion of the ligand molecule as a strategy for modulating binding affinity: Further studies involving complexes of jacalin with beta-substituted disaccharides.
IUBMB Life, 69, 2017
|
|
5JBS
 
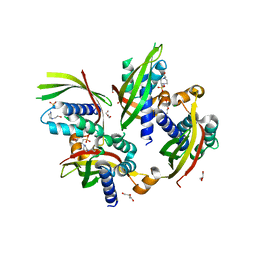 | | Conformational changes during monomer-to-dimer transition of Brucella suis VirB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arya, T, Sharifahmadian, M, Sygusch, J, Baron, B. | | Deposit date: | 2016-04-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR analyses, X-ray crystallography and small-molecule probing reveal conformational shifts during monomer-to-dimer transition of Brucella suis VirB8
To Be Published
|
|
7RNJ
 
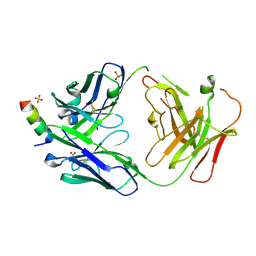 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
5ITX
 
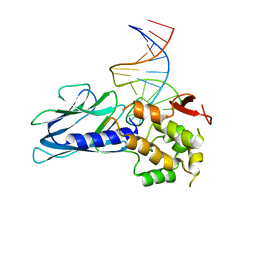 | | Crystal Structure of Human NEIL1(P2G R242K) bound to duplex DNA containing Thymine Glycol | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IU9
 
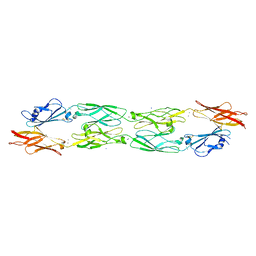 | |
5IUX
 
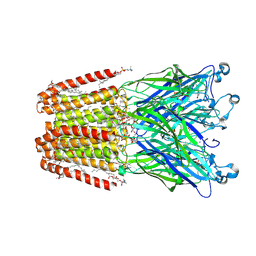 | | GLIC-V135C bimane labelled X-ray structure | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, ACETATE ION, ... | | Authors: | Fourati, Z, Menny, A, Delarue, M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a pre-active conformation of a pentameric channel receptor.
Elife, 6, 2017
|
|
5J5G
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 6-(4-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
3ZNY
 
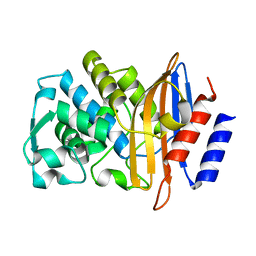 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX- M-96, a natural D240G mutant derived from CTX-M-12 | | Descriptor: | CTX-M-12A ENZYME | | Authors: | Power, P, Herman, R, Bouillenne, F, Ghiglione, B, Rodriguez, M.M, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2013-02-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Insights Into the "Ceftazidimase" Behavior of the Extended-Spectrum Beta-Lactamase Ctx-M-96.
Biochemistry, 54, 2015
|
|
6Z1B
 
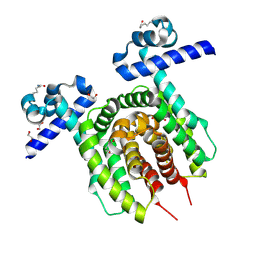 | |
7RL1
 
 | | AAVrh.10-7x capsid | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
7XE4
 
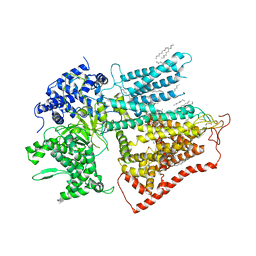 | | structure of a membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
3ZPY
 
 | | Crystal structure of the marine PL7 alginate lyase AlyA1 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, SODIUM ION | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
3ZRV
 
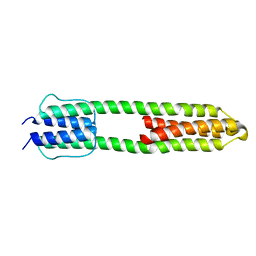 | | The high resolution structure of a dimeric Hamp-Dhp fusion displays asymmetry - A291F mutant | | Descriptor: | HAMP, OSMOLARITY SENSOR PROTEIN ENVZ | | Authors: | Zeth, K, Hulko, M, Ferris, H.U, Martin, J. | | Deposit date: | 2011-06-20 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Regulation of Receptor Histidine Kinases.
Structure, 20, 2012
|
|
7RF6
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF4
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
