3RHK
 
 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
3RHX
 
 | | Crystal structure of the catalytic domain of FGFR1 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, 1,2-ETHANEDIOL, Basic fibroblast growth factor receptor 1, ... | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
6K37
 
 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with NAD+ and the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | (3R)-3-[(1R)-1-azanylethyl]nonanedioic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Nishiyama, M. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
3O6I
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-[({3-tert-butyl-4-[(methylamino)methyl]-1H-pyrazol-1-yl}acetyl)amino]-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3O74
 
 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
6KMD
 
 | | Crystal structure of SeMet-phytochromobilin synthase from tomato in complex with biliverdin | | Descriptor: | BILIVERDINE IX ALPHA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sugishima, M, Wada, K, Fukuyama, K. | | Deposit date: | 2019-07-31 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phytochromobilin synthase in complex with biliverdin IX alpha , a key enzyme in the biosynthesis of phytochrome.
J.Biol.Chem., 295, 2020
|
|
6K0V
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
3NST
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans | | Descriptor: | FE (II) ION, GLYCEROL, Gentisate 1,2-dioxygenase | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
3NSL
 
 | |
6K2V
 
 | |
6JU1
 
 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
3NWS
 
 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
3NWG
 
 | | The crystal structure of a microcomparments protein from Desulfitobacterium hafniense DCB | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Fan, Y, Volkart, L, Gu, M, Axen, S, Greenleaf, W.B, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-09 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a PduT homolog from a novel bacterial microcompartment
To be Published
|
|
6JY9
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6KGB
 
 | |
3O1K
 
 | | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Dihydroneopterin aldolase FolB, putative | | Authors: | Nocek, B, Zhou, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3O45
 
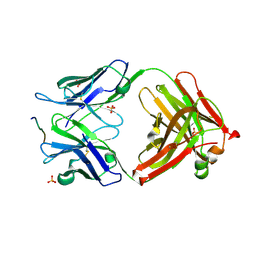 | | Crystal Structure of 101F Fab Bound to 17-mer Peptide Epitope | | Descriptor: | Fusion glycoprotein F1, Mouse monoclonal antibody 101F 101F Fab heavy chain, Mouse monoclonal antibody 101F 101F Fab light chain, ... | | Authors: | McLellan, J.S, Chen, M, Chang, J.S, Yang, Y, Kim, A, Graham, B.S, Kwong, P.D. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Structure of a Major Antigenic Site on the Respiratory Syncytial Virus Fusion Glycoprotein in Complex with Neutralizing Antibody 101F.
J.Virol., 84, 2010
|
|
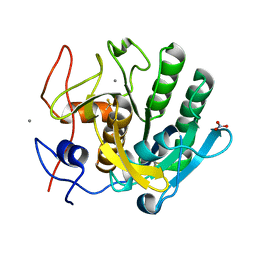
6K2T
 
 | |
3O4D
 
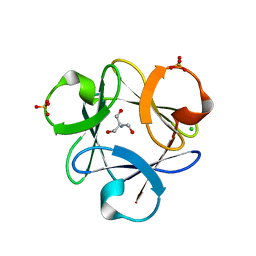 | |
6K60
 
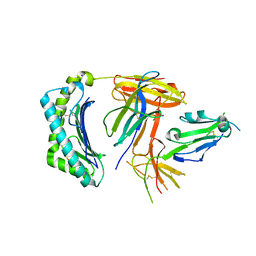 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
3O8G
 
 | | EthR from Mycobacterium tuberculosis in complex with compound BDM14801 | | Descriptor: | 1-(azidoacetyl)-4-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)piperidine, TRANSCRIPTIONAL REGULATORY REPRESSOR PROTEIN (TETR-FAMILY) ETHR | | Authors: | Willand, N, Desroses, M, Toto, P, Diri, B, Lens, Z, Villeret, V, Rucktooa, P, Locht, C, Baulard, A, Deprez, B. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring drug target flexibility using in situ click chemistry: application to a mycobacterial transcriptional regulator.
Acs Chem.Biol., 5, 2010
|
|
6KNF
 
 | | CryoEM map and model of Nitrite Reductase at pH 6.2 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
3O9N
 
 | | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Singh, A, Kumar, S, Sinha, M, Sharma, S, Singh, T.P. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a new form of xylanase-A-amylase inhibitor protein(XAIP-III) at 2.4 A resolution
To be Published
|
|
3RT9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A | | Descriptor: | COENZYME A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RF0
 
 | | Crystal Structure of Exopolyphosphatase from Yersinia pestis | | Descriptor: | Exopolyphosphatase, FORMIC ACID, NITRATE ION | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Exopolyphosphatase from Yersinia pestis
To be Published
|
|
