6IQT
 
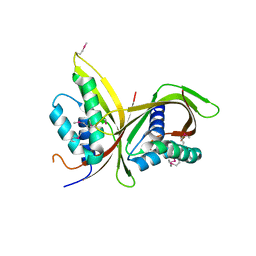 | | Crystal Structure of CagV, a VirB8 homolog of T4SS from Helicobacter pylori Strain 26695 | | Descriptor: | Cag pathogenicity island protein (Cag10) | | Authors: | Wu, X, Zhao, Y, Sun, L, Jiang, M, Wang, Q, Wang, Q, Yang, W, Wu, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal structure of CagV, the Helicobacter pylori homologue of the T4SS protein VirB8.
Febs J., 286, 2019
|
|
3REV
 
 | | Crystal structure of human alloreactive tcr nb20 | | Descriptor: | 1,2-ETHANEDIOL, TCR NB20 ALPHA CHAIN, TCR NB20 BETA CHAIN | | Authors: | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | Deposit date: | 2011-04-05 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RF1
 
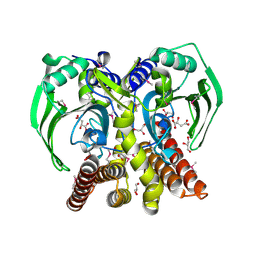 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Glycyl-tRNA synthetase alpha subunit | | Authors: | Tan, K, Zhang, R, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-05 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
6ISG
 
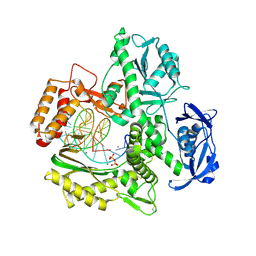 | | Structure of 9N-I DNA polymerase incorporation with dG in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
3R5P
 
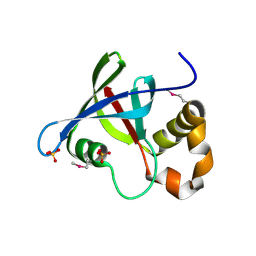 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3RLB
 
 | | Crystal structure at 2.0 A of the S-component for thiamin from an ECF-type ABC transporter | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ThiT, nonyl beta-D-glucopyranoside | | Authors: | Erkens, G.B, Berntsson, R.P.-A, Fulyani, F, Majsnerowska, M, Vujicic-Zagar, A, ter Beek, J, Poolman, B, Slotboom, D.J. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of modularity in ECF-type ABC transporters.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6I1O
 
 | | Fab fragment of an antibody selective for wild-type alpha-1-antitrypsin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FAB 2H2 heavy chain, FAB 2H2 light chain, ... | | Authors: | Laffranchi, M, Elliston, E.L.K, Miranda, E, Perez, J, Fra, A, Lomas, D.A, Irving, J.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Intrahepatic heteropolymerization of M and Z alpha-1-antitrypsin.
JCI Insight, 5, 2020
|
|
3R75
 
 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 in complex with benzoate, pyruvate, glutamine and contaminating Zn2+ | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3ROE
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Thymidine | | Descriptor: | Apolipoprotein A-I-binding protein, THYMIDINE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-25 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6I69
 
 | |
3RQX
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRB
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6JY8
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
5TJK
 
 | | Crystal structure of GTA + A trisaccharide (native) | | Descriptor: | GLYCEROL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
3R4X
 
 | | Crystal structure of bovine lactoperoxidase complexed with pyrazine-2-carboxamide at 2 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of bovine lactoperoxidase complexed with Pyrazine-2-carboxamide at 2 A resolution
To be Published
|
|
3R5W
 
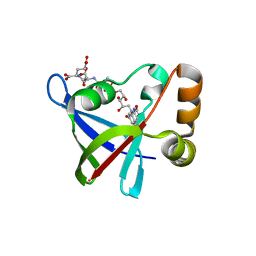 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3RE1
 
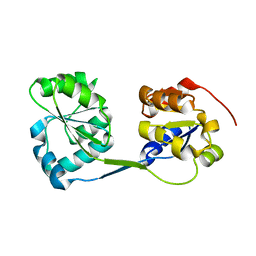 | |
6K5W
 
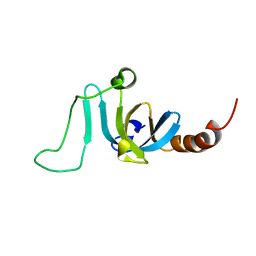 | | Solution structure of the chromodomain of yeast Eaf3 | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2019-05-31 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eaf3 chromodomain acts as a pH sensor for gene expression by altering its binding affinity for histone methylated-lysine residues.
Biosci.Rep., 40, 2020
|
|
6K5Q
 
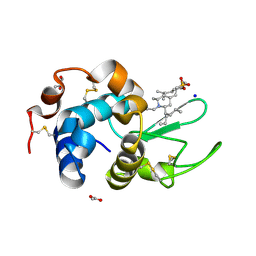 | | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia | | Descriptor: | ACETATE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Kanal Elamparithi, B, Ankur, T, Sivakumar, M, Gunasekaran, K. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia
To Be Published
|
|
6K6V
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-06-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6K7K
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ADP-Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7X
 
 | | Human MCU-EMRE complex | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
6JY7
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
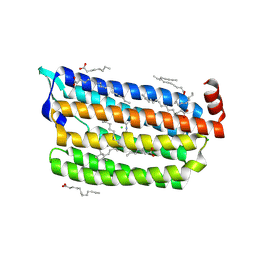 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
