6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
3RC3
 
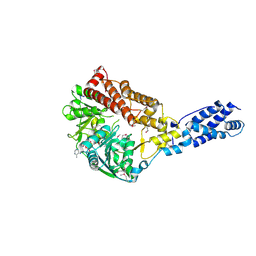 | | Human Mitochondrial Helicase Suv3 | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, AZIDE ION, ... | | Authors: | Dauter, Z, Jedrzejczak, R, Dauter, M, Szczesny, R, Stepien, P. | | Deposit date: | 2011-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3R2M
 
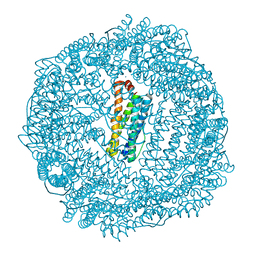 | | 1.8A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2T
 
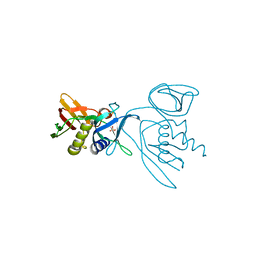 | | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325. | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325.
TO BE PUBLISHED
|
|
3R45
 
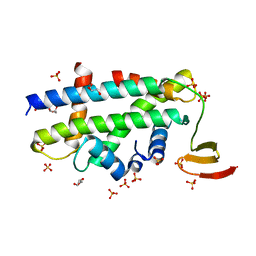 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
6JUX
 
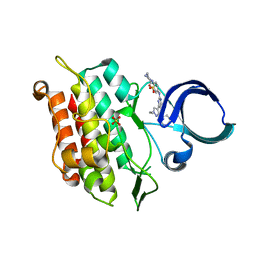 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-71807 | | Descriptor: | 4-(1-ethyl-3-pyridin-3-yl-pyrazol-4-yl)-~{N}-(4-piperazin-1-ylphenyl)pyrimidin-2-amine, Activin receptor type-1, SULFATE ION | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Activin Receptor-Like Kinase 2 (R206H) Inhibition by Bis-heteroaryl Pyrazole-Based Inhibitors for the Treatment of Fibrodysplasia Ossificans Progressiva Identified by the Integration of Ligand-Based and Structure-Based Drug Design Approaches.
Acs Omega, 5, 2020
|
|
6KEX
 
 | |
6KGA
 
 | |
6KHJ
 
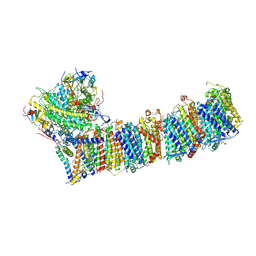 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
6IAR
 
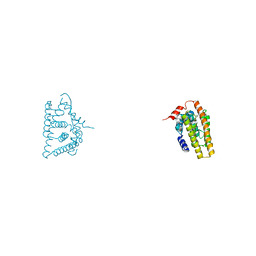 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | Descriptor: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | Authors: | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
6IBG
 
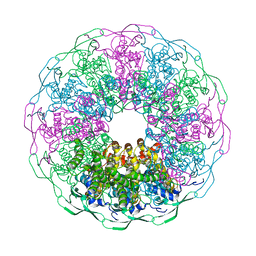 | | Bacteriophage G20c portal protein crystal structure for construct with intact N-terminus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Portal protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3RSS
 
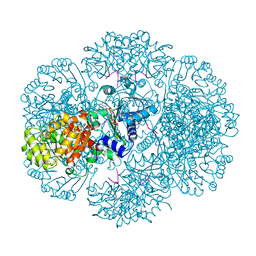 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTC
 
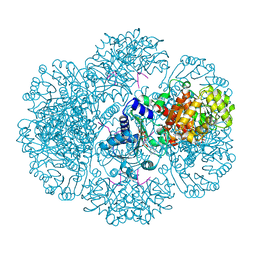 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NAD and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6I6W
 
 | |
3QSL
 
 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
3QAP
 
 | | Crystal structure of Natronomonas pharaonis sensory rhodopsin II in the ground state | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, EICOSANE, RETINAL, ... | | Authors: | Gushchin, I, Reshetnyak, A, Borshchevskiy, V, Ishchenko, A, Round, E, Grudinin, S, Engelhard, M, Buldt, G, Gordeliy, V. | | Deposit date: | 2011-01-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active State of Sensory Rhodopsin II: Structural Determinants for Signal Transfer and Proton Pumping.
J.Mol.Biol., 412, 2011
|
|
3QCZ
 
 | | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase with Mn, AMPPNP and L-Glutamate bound | | Descriptor: | Dihydrofolate synthase / folylpolyglutamate synthase, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase with Mn, AMPPNP and L-Glutamate bound
TO BE PUBLISHED
|
|
6IH1
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-GMP bound form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
3QGC
 
 | | Crystal structure of FBF-2 R288Y mutant in complex with gld-1 FBEa A7U mutant | | Descriptor: | 5'-R(*UP*GP*UP*GP*CP*CP*UP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Qiu, C, Koh, Y.Y, Opperman, L, Gross, L, Hall, T.M.T, Wickens, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stacking interactions in PUF-RNA complexes.
Rna, 17, 2011
|
|
3QGZ
 
 | | Re-investigated high resolution crystal structure of histidine triad nucleotide-binding protein 1 (HINT1) from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A, Nawrot, B, Stec, W.J. | | Deposit date: | 2011-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution X-ray crystal structure of rabbit histidine triad nucleotide-binding protein 1 (rHINT1) - adenosine complex at 1.10A resolution
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6IJO
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6IFQ
 
 | |
6INB
 
 | |
3QND
 
 | |
3QO4
 
 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
