6ZEJ
 
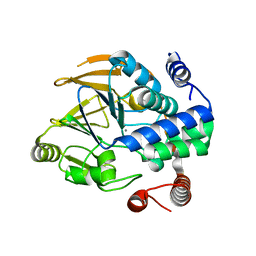 | | Structure of PP1-Phactr1 chimera [PP1(7-304) + linker (SGSGS) + Phactr1(526-580)] | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6Z3B
 
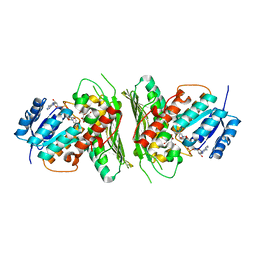 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
4NG1
 
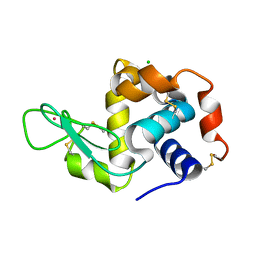 | | Previously de-ionized HEW lysozyme batch crystallized in 1.9 M CsCl | | Descriptor: | CESIUM ION, CHLORIDE ION, Lysozyme C | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NGI
 
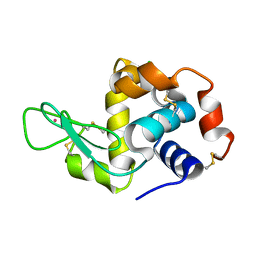 | | Previously de-ionized HEW lysozyme crystallized in 1.0 M RbCl and collected at 125K | | Descriptor: | CHLORIDE ION, Lysozyme C, RUBIDIUM ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NFV
 
 | | Previously de-ionized HEW lysozyme batch crystallized in 1.1 M MnCl2 | | Descriptor: | CHLORIDE ION, Lysozyme C, MANGANESE (II) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6ZF9
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4
To Be Published
|
|
6YTL
 
 | |
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | Deposit date: | 2020-06-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
6YU7
 
 | | Crystal structure of MhsT in complex with L-tyrosine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6ZI1
 
 | | Crystal structure of the isolated H. influenzae VapD toxin (D7N mutant) | | Descriptor: | Endoribonuclease VapD | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
6YTV
 
 | |
6YU2
 
 | | Crystal structure of MhsT in complex with L-isoleucine | | Descriptor: | ISOLEUCINE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6Z08
 
 | | Crystal structure of cAMP-dependent protein kinase A (CHO PKA) in complex with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Oebbeke, M, Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2020-05-08 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZAU
 
 | | Damage-free nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZEA
 
 | | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{R})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, (1~{S})-1-methyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, 1,2-ETHANEDIOL, ... | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Strictosidine Synthase from Catharanthus roseus in complex with racemic 1-methyl-2,3,4,9-tetrahydro-1H-beta-carboline
To Be Published
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6YU3
 
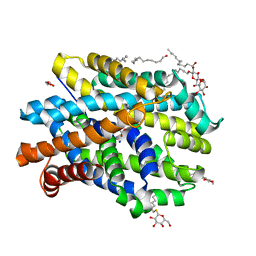 | | Crystal structure of MhsT in complex with L-phenylalanine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, GLYCEROL, PHENYLALANINE, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
6YWO
 
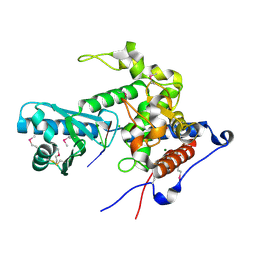 | | CutA in complex with A3 RNA | | Descriptor: | CHLORIDE ION, CutA, MAGNESIUM ION, ... | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
3DBS
 
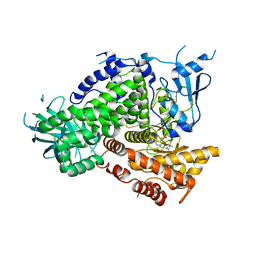 | | Structure of PI3K gamma in complex with GDC0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Wiesmann, C, Ultsch, M. | | Deposit date: | 2008-06-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer
J.Med.Chem., 51, 2008
|
|
6Z1Q
 
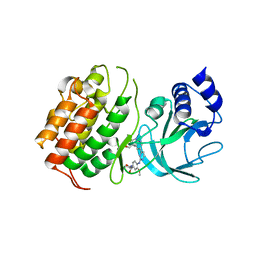 | | MAP3K14 (NIK) in complex with DesF-3R/4076 | | Descriptor: | DesF-3R/4076, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Jacoby, E, van Vlijmen, H, Querolle, O, Stansfield, I, Meerpoel, L, Versele, M, Hynd, G, Attar, R. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | FEP+ calculations predict a stereochemical SAR switch for first-in-class indoline NIK inhibitors for multiple myeloma
Future Drug Discov, 2, 2020
|
|
6Z20
 
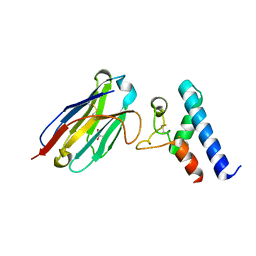 | | Structure of the EC2 domain of CD9 in complex with nanobody 4C8 | | Descriptor: | CD9 antigen, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oosterheert, W, Manshande, J, Pearce, N.M, Lutz, M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
4NGL
 
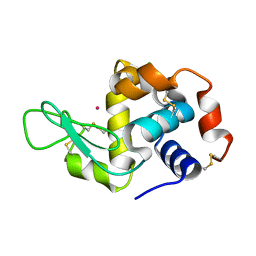 | | Previously de-ionized HEW lysozyme batch crystallized in 0.6 M CoCl2 | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Lysozyme C | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6Z7U
 
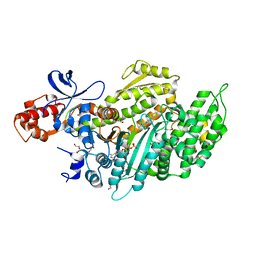 | | Myosin-II motor domain complexed with blebbistatin in a new ADP-release conformation | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ewert, W, Preller, M. | | Deposit date: | 2020-06-01 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and Computational Insights into a Blebbistatin-Bound Myosin•ADP Complex with Characteristics of an ADP-Release Conformation along the Two-Step Myosin Power Stoke.
Int J Mol Sci, 21, 2020
|
|
6ZAS
 
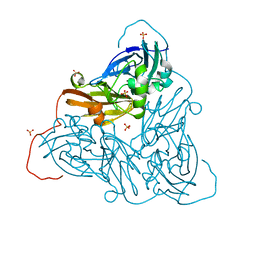 | | Damage-free as-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2020-06-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
