7Q0F
 
 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7PZY
 
 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q08
 
 | | Structure of Candida albicans 80S ribosome in complex with cycloheximide | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7UFS
 
 | | Cryo-EM Structure of Bl_Man38B at 3.4 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of Bl_Man38B at 3.4 A
Nat.Chem.Biol., 2022
|
|
8WKZ
 
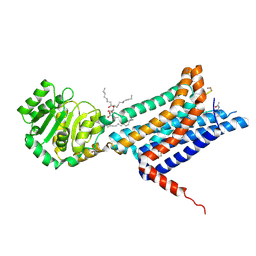 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
7Q0R
 
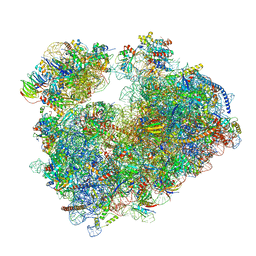 | | Structure of the Candida albicans 80S ribosome in complex with blasticidin s | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q34
 
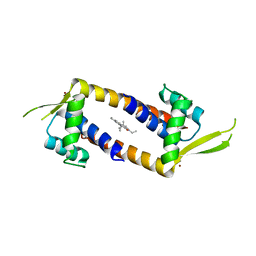 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex squaraine dye | | Descriptor: | 2,4-bis[(E)-(1-ethyl-3,3-dimethyl-indol-2-ylidene)methyl]cyclobutane-1,3-dione, Helix-turn-helix transcriptional regulator, NICKEL (II) ION | | Authors: | Liutkus, M, Mejias, S.H, Barolo, C, Cortajarena, A.L. | | Deposit date: | 2021-10-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Designing Artificial Fluorescent Proteins: Squaraine-LmrR Biophosphors for High Performance Deep-Red Biohybrid Light-Emitting Diodes
Adv Funct Mater, 32, 2022
|
|
7UVE
 
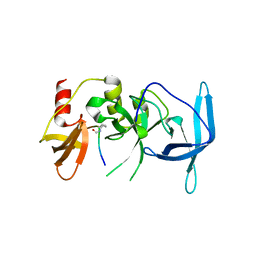 | | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac | | Descriptor: | Histone-lysine N-methyltransferase eggless, peptide H3K9me2K14ac | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-01 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain with peptide H3K9me2K14ac
To Be Published
|
|
7UW8
 
 | | Drosophila melanogaster setdb1-tuor domain | | Descriptor: | Histone-lysine N-methyltransferase eggless | | Authors: | Zhou, M, Dong, A, Liu, K, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Drosophila melanogaster setdb1-tuor domain
To Be Published
|
|
4TNM
 
 | |
7UPS
 
 | |
8XI9
 
 | | Crystal structure of FRB-FKBP fusion protein in complex with rapamycin | | Descriptor: | FRB-FKBP fusion protein, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Inobe, T, Sakaguchi, R, Obita, T, Mukaiyama, A, Yokoyama, T, Mizuguchi, M, Akiyama, S. | | Deposit date: | 2023-12-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into rapamycin-induced oligomerization of a FRB-FKBP fusion protein.
Febs Lett., 598, 2024
|
|
7UFT
 
 | | Cryo-EM Structure of Bl_Man38C at 2.9 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure of Bl_Man38C at 2.9 A
Nat.Chem.Biol., 2022
|
|
8WGO
 
 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of PneA and GTPrS. | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
1ESM
 
 | | STRUCTURAL BASIS FOR THE FEEDBACK REGULATION OF ESCHERICHIA COLI PANTOTHENATE KINASE BY COENZYME A | | Descriptor: | COENZYME A, PANTOTHENATE KINASE | | Authors: | Yun, M, Park, C.G, Kim, J.Y, Rock, C.O, Jackowski, S, Park, H.W. | | Deposit date: | 2000-04-10 | | Release date: | 2000-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the feedback regulation of Escherichia coli pantothenate kinase by coenzyme A.
J.Biol.Chem., 275, 2000
|
|
7UPV
 
 | |
7UPZ
 
 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
1EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE) | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
1EFO
 
 | | CRYSTAL STRUCTURE OF AN ADENINE BULGE IN THE RNA CHAIN OF A DNA/RNA HYBRID, D(CTCCTCTTC)/R(GAAGAGAGAG) | | Descriptor: | DNA (5'-D(*CP*TP*CP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3') | | Authors: | Sudarsanakumar, C, Xiong, Y, Sundaralingam, M. | | Deposit date: | 2000-02-09 | | Release date: | 2000-05-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine bulge in the RNA chain of a DNA.RNA hybrid, d(CTCCTCTTC).r(gaagagagag).
J.Mol.Biol., 299, 2000
|
|
6JIB
 
 | | Human MTHFD2 in complex with DS44960156 | | Descriptor: | 4-(5-oxo-1,5-dihydro-2H-[1]benzopyrano[3,4-c]pyridine-3(4H)-carbonyl)benzoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
1EVP
 
 | |
6JNF
 
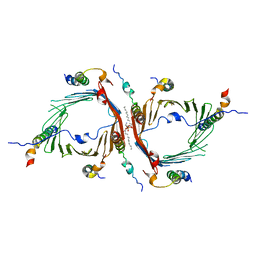 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
1EIY
 
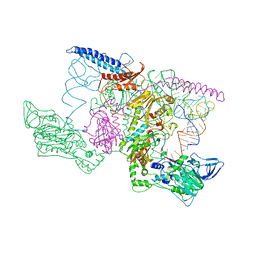 | | THE CRYSTAL STRUCTURE OF PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH COGNATE TRNAPHE | | Descriptor: | PHENYLALANYL-TRNA SYNTHETASE, TRNA(PHE) | | Authors: | Goldgur, Y, Mosyak, L, Reshetnikova, L, Ankilova, V, Safro, M. | | Deposit date: | 2000-02-29 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe.
Structure, 5, 1997
|
|
1E5U
 
 | | NMR Representative Structure of Intimin-190 (Int190) from Enteropathogenic E. coli | | Descriptor: | INTIMIN | | Authors: | Prasannan, S, Matthews, S.J, Batchelor, M, Daniell, S, Reece, S, Frankel, G, Dougan, G, Connerton, I, Bloomberg, G. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Recognition of the Translocated Intimin Receptor (Tir) by Intimin from Enteropathogenic E. Coli
Embo J., 19, 2000
|
|
6JQF
 
 | |
