1S4S
 
 | | Reaction Intermediate in the Photocycle of PYP, intermediate occupied between 100 micro-seconds to 5 milli-seconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Schmidt, M, Pahl, R, Srajer, V, Anderson, S, Ren, Z, Ihee, H, Rajagopal, S, Moffat, K. | | Deposit date: | 2004-01-17 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3R36
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73Q complexed with 4-hydroxybenzoic acid | | Descriptor: | 4-hydroxybenzoyl-CoA thioesterase, P-HYDROXYBENZOIC ACID | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
5EAH
 
 | | Saccharomyces cerevisiae CYP51 complexed with the plant pathogen inhibitor Difenoconazole | | Descriptor: | 1-[[(2~{R},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{R},4~{S})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, 1-[[(2~{S},4~{R})-2-[2-chloranyl-4-(4-chloranylphenoxy)phenyl]-4-methyl-1,3-dioxolan-2-yl]methyl]-1,2,4-triazole, ... | | Authors: | Tyndall, J.D.A, Sabherwal, M, Keniya, M.V, Wilson, R.K, Woods, M.V, Monk, B.C. | | Deposit date: | 2015-10-16 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural and Functional Elucidation of Yeast Lanosterol 14 alpha-Demethylase in Complex with Agrochemical Antifungals.
PLoS ONE, 11, 2016
|
|
7F3E
 
 | | Cryo-EM structure of the minimal protein-only RNase P from Aquifex aeolicus | | Descriptor: | RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Adachi, N, Kawasaki, M, Moriya, T, Numata, T, Senda, T, Kakuta, Y. | | Deposit date: | 2021-06-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Minimal protein-only RNase P structure reveals insights into tRNA precursor recognition and catalysis.
J.Biol.Chem., 297, 2021
|
|
5DW7
 
 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
5OFU
 
 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in T-state. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
7EQD
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Yu, L.-J, Hall, M, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-05-01 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM Structure of the Photosynthetic LH1-RC Complex from Rhodospirillum rubrum .
Biochemistry, 2021
|
|
5OH5
 
 | | Legionella pneumophila RidL N-terminal retromer binding domain | | Descriptor: | RidL | | Authors: | Baerlocher, K, Hutter, C.A.J, Swart, A.L, Steiner, B, Welin, A, Hohl, M, Letourneur, F, Seeger, M.A, Hilbi, H. | | Deposit date: | 2017-07-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into Legionella RidL-Vps29 retromer subunit interaction reveal displacement of the regulator TBC1D5.
Nat Commun, 8, 2017
|
|
5OBW
 
 | | Mycoplasma genitalium DnaK-NBD in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5DZA
 
 | | Streptococcus agalactiae AgI/II polypeptide BspA C terminal domain (WT) | | Descriptor: | 1,2-ETHANEDIOL, BspA, DI(HYDROXYETHYL)ETHER | | Authors: | Rego, S, Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-06-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae.
J.Biol.Chem., 291, 2016
|
|
3ATJ
 
 | | HEME LIGAND MUTANT OF RECOMBINANT HORSERADISH PEROXIDASE IN COMPLEX WITH BENZHYDROXAMIC ACID | | Descriptor: | BENZHYDROXAMIC ACID, CALCIUM ION, PROTEIN (HORSERADISH PEROXIDASE C1A), ... | | Authors: | Meno, K, White, C.G, Smith, A.T, Gajhede, M. | | Deposit date: | 1998-12-16 | | Release date: | 1999-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Catalytical Implications of a F221M Mutation in the Proximal Pocket of Horseradish Peroxidase C (HRP C)
To be Published
|
|
5ANB
 
 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
7F73
 
 | | Cryo-EM structure of human TMEM120B | | Descriptor: | MCherry fluorescent protein,Transmembrane protein 120B | | Authors: | Ke, M, Wu, J, Yan, Z. | | Deposit date: | 2021-06-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human TMEM120A and TMEM120B.
Cell Discov, 7, 2021
|
|
5AV1
 
 | |
5NUH
 
 | |
1SJX
 
 | | Three-Dimensional Structure of a Llama VHH Domain OE7 binding the cell wall protein Malf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, immunoglobulin VH domain | | Authors: | Dolk, E, van der Vaart, M, Hulsik, D.L, Vriend, G, de Haard, H, Spinelli, S, Cambillau, C, Frenken, L, Verrips, T. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo.
Appl.Environ.Microbiol., 71, 2005
|
|
3R3C
 
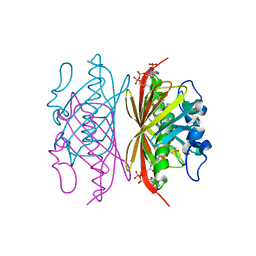 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant H64A complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA thioesterase | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
5NUV
 
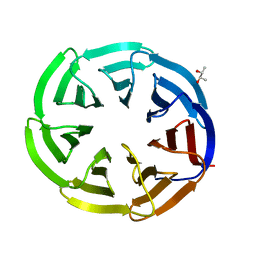 | |
5BNF
 
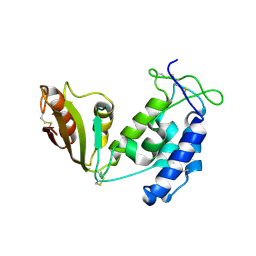 | | Apo structure of porcine CD38 | | Descriptor: | Uncharacterized protein | | Authors: | Ting, K.Y, Leung, C.P.F, Graeff, R.M, Lee, H.C, Hao, Q, Kotaka, M. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porcine CD38 exhibits prominent secondary NAD(+) cyclase activity.
Protein Sci., 25, 2016
|
|
5NV3
 
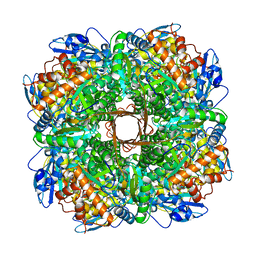 | | Structure of Rubisco from Rhodobacter sphaeroides in complex with CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Bracher, A, Milicic, G, Ciniawsky, S, Wendler, P, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-26 | | Last modified: | 2017-09-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Enzyme Repair by the AAA(+) Chaperone Rubisco Activase.
Mol. Cell, 67, 2017
|
|
5AWJ
 
 | | Crystal structure of VDR-LBD/partial agonist complex: 22S-hexyl analogue | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)nonan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor,Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Inaba, Y, Nakabayashi, M, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2015-07-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Bioorg.Med.Chem., 23, 2015
|
|
5BR2
 
 | | Structure of bacteriorhodopsin crystallized from ND-MSP1 | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Nikolaev, M, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-05-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integral Membrane Proteins Can Be Crystallized Directly from Nanodiscs
Cryst.Growth Des., 17, 2017
|
|
5AX2
 
 | | Crystal structure of S.cerevisiae Kti11p | | Descriptor: | CADMIUM ION, Diphthamide biosynthesis protein 3 | | Authors: | Kumar, A, Nagarathinam, K, Tanabe, M, Balbach, J. | | Deposit date: | 2015-07-13 | | Release date: | 2016-07-20 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hyperbolic Pressure-Temperature Phase Diagram of the Zinc-Finger Protein apoKti11 Detected by NMR Spectroscopy.
J Phys Chem B, 123, 2019
|
|
7EQ9
 
 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | Descriptor: | TIP60 | | Authors: | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | Deposit date: | 2021-04-30 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
5ANC
 
 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
