1HSN
 
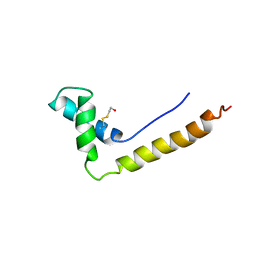 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
2L44
 
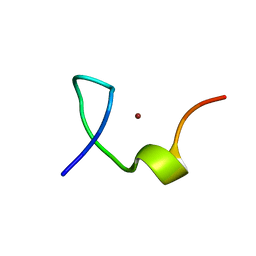 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
3QY5
 
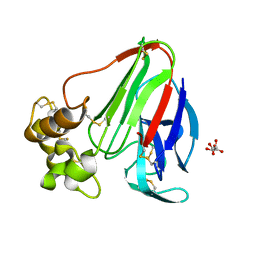 | |
2MXC
 
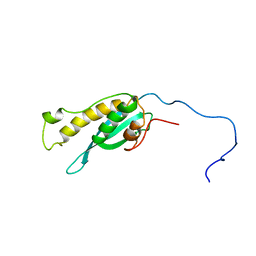 | | Solution structure of the full length sorting nexin 3 | | Descriptor: | Sorting nexin-3 | | Authors: | Lenoir, M.M.L, Rajesh, S.S.R, Gruenberg, J.J.G, Overduin, M.M.O, Kaur, J.J.K. | | Deposit date: | 2014-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of conserved phosphoinositide binding pocket regulates sorting nexin membrane targeting.
Nat Commun, 9, 2018
|
|
3QY4
 
 | |
2V9U
 
 | | Rim domain of main porin from Mycobacteria smegmatis | | Descriptor: | MSPA | | Authors: | Grueninger, D, Ziegler, M.O.P, Koetter, J.W.A, Treiber, N, Schulze, M.-S, Schulz, G.E. | | Deposit date: | 2007-08-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Designed Protein-Protein Association.
Science, 319, 2008
|
|
2VL8
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP, CASTANOSPERMINE AND CALCIUM ION | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, CYTOTOXIN L, ... | | Authors: | Jank, T, Ziegler, M.O.P, Schulz, G.E, Aktories, K. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inhibition of the Glucosyltransferase Activity of Clostridial Rho/Ras-Glucosylating Toxins by Castanospermine.
FEBS Lett., 582, 2008
|
|
2VK9
 
 | |
2VKH
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP-GLC AND CALCIUM ION | | Descriptor: | CALCIUM ION, CYTOTOXIN L, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ziegler, M.O.P, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes and Reaction of Clostridial Glycosylating Toxins.
J.Mol.Biol., 377, 2008
|
|
2VKD
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP-GLC AND MANGANESE ION | | Descriptor: | CYTOTOXIN L, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ziegler, M.O.P, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Conformational Changes and Reaction of Clostridial Glycosylating Toxins.
J.Mol.Biol., 377, 2008
|
|
2RHB
 
 | | Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Palaninathan, S, Bhardwaj, K, Alcantara, J.M.O, Guarino, L, Yi, L.L, Kao, C.C, Sacchettini, J. | | Deposit date: | 2007-10-08 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.
J.Biol.Chem., 283, 2008
|
|
6EYO
 
 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
5LGJ
 
 | |
5LGK
 
 | | Crystal structure of the human IgE-Fc bound to its B cell receptor derCD23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig epsilon chain C region, Low affinity immunoglobulin epsilon Fc receptor, ... | | Authors: | Dhaliwal, B, Pang, M.O.Y, Sutton, B.J. | | Deposit date: | 2016-07-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | IgE binds asymmetrically to its B cell receptor CD23.
Sci Rep, 7, 2017
|
|
