3HHB
 
 | |
3I2N
 
 | | Crystal Structure of WD40 repeats protein WDR92 | | Descriptor: | WD repeat-containing protein 92 | | Authors: | Amaya, M.F, Li, Z, He, H, Seitova, A, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3I91
 
 | | Crystal structure of human chromobox homolog 8 (CBX8) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 8, H3K9 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
1DK4
 
 | | CRYSTAL STRUCTURE OF MJ0109 GENE PRODUCT INOSITOL MONOPHOSPHATASE | | Descriptor: | INOSITOL MONOPHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Stec, B, Yang, H, Johnson, K.A, Chen, L, Roberts, M.F. | | Deposit date: | 1999-12-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MJ0109 is an enzyme that is both an inositol monophosphatase and the 'missing' archaeal fructose-1,6-bisphosphatase.
Nat.Struct.Biol., 7, 2000
|
|
3I8Z
 
 | | Crystal structure of human chromobox homolog 4 (CBX4) | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Amaya, M.F, Zhihong, L, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of human chromobox homolog 4 (CBX4)
To be Published
|
|
3I90
 
 | | Crystal structure of human chromobox homolog 6 (CBX6) with H3K27 peptide | | Descriptor: | Chromobox protein homolog 6, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
1B0Q
 
 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3FDT
 
 | | Crystal structure of the complex of human chromobox homolog 5 (CBX5) with H3K9(me)3 peptide | | Descriptor: | Chromobox protein homolog 5, H3K9(me)3 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
3H91
 
 | | Crystal structure of the complex of human chromobox homolog 2 (CBX2) and H3K27 peptide | | Descriptor: | Chromobox protein homolog 2, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
3HM5
 
 | | SANT domain of human DNA methyltransferase 1 associated protein 1 | | Descriptor: | CALCIUM ION, DNA methyltransferase 1-associated protein 1, UNKNOWN ATOM OR ION | | Authors: | Dombrovski, L, Tempel, W, Amaya, M.F, Tong, Y, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Park, H, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SANT domain of human DNA methyltransferase 1 associated protein 1
To be Published
|
|
3HXN
 
 | |
3I3C
 
 | | Crystal Structural of CBX5 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 5, SODIUM ION | | Authors: | Amaya, M.F, Li, Z, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structural of CBX5 Chromo Shadow Domain
To be Published
|
|
3IVQ
 
 | | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases: SPOPMATH-CiSBC2 | | Descriptor: | CiSBC2, Speckle-type POZ protein | | Authors: | Schulman, B.A, Miller, D.J, Calabrese, M.F, Seyedin, S. | | Deposit date: | 2009-09-01 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases.
Mol.Cell, 36, 2009
|
|
3GV6
 
 | | Crystal Structure of human chromobox homolog 6 (CBX6) with H3K9 peptide | | Descriptor: | Chromobox protein homolog 6, Histone H3K9me3 peptide | | Authors: | Dong, A, Amaya, M.F, Li, Z, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
3IHO
 
 | | The C-terminal glycosylase domain of human MBD4 | | Descriptor: | Methyl-CpG-binding domain protein 4 | | Authors: | Amaya, M.F, Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-terminal glycosylase domain of human MBD4
To be Published
|
|
1E3H
 
 | | SeMet derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | GUANOSINE PENTAPHOSPHATE SYNTHETASE, SULFATE ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-15 | | Release date: | 2000-11-05 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1DSQ
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
3GFC
 
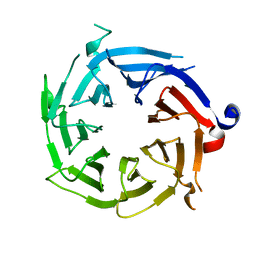 | | Crystal Structure of Histone-binding protein RBBP4 | | Descriptor: | Histone-binding protein RBBP4 | | Authors: | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
1E15
 
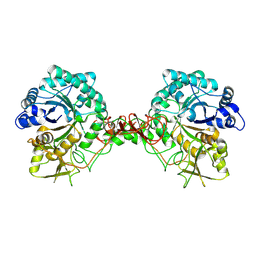 | | Chitinase B from Serratia Marcescens | | Descriptor: | CHITINASE B | | Authors: | Van Aalten, D.M.F, Synstad, B, Brurberg, M.B, Hough, E, Riise, B.W, Eijsink, V.G.H, Wierenga, R.K. | | Deposit date: | 2000-04-18 | | Release date: | 2000-08-18 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Two-Domain Chitotriosidase from Serratia Marcescens at 1.9 Angstrom Resoltuion
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E2X
 
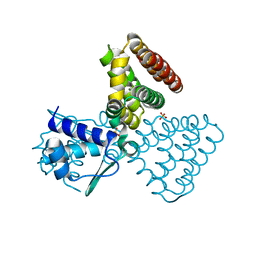 | | FadR, fatty acid responsive transcription factor from E. coli | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J, Wierenga, R.K. | | Deposit date: | 2000-05-30 | | Release date: | 2000-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fadr, a Fatty Acid-Responsive Transcription Factor with a Novel Acyl Coenzyme A-Binding Fold
Embo J., 19, 2000
|
|
1E3P
 
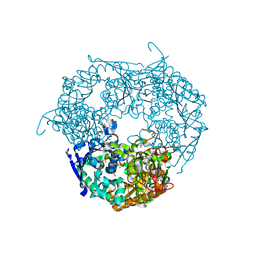 | | tungstate derivative of Streptomyces antibioticus PNPase/GPSI enzyme | | Descriptor: | Polyribonucleotide nucleotidyltransferase, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Symmons, M.F, Jones, G.H, Luisi, B.F. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Duplicated Fold is the Structural Basis for Polynucleotide Phosphorylase Catalytic Activity, Processivity, and Regulation
Structure, 8, 2000
|
|
1DZT
 
 | | RMLC FROM SALMONELLA TYPHIMURIUM | | Descriptor: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
3GNI
 
 | | Structure of STRAD and MO25 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Protein Mo25, ... | | Authors: | Zeqiraj, E, Goldie, S, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2009-03-17 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor
Plos Biol., 7, 2009
|
|
1A6X
 
 | | STRUCTURE OF THE APO-BIOTIN CARBOXYL CARRIER PROTEIN (APO-BCCP87) OF ESCHERICHIA COLI ACETYL-COA CARBOXYLASE, NMR, 49 STRUCTURES | | Descriptor: | APO-BIOTIN CARBOXYL CARRIER PROTEIN OF ACETYL-COA CARBOXYLASE | | Authors: | Yao, X, Wei, D, Soden Junior, C, Summers, M.F, Beckett, D. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the carboxy-terminal fragment of the apo-biotin carboxyl carrier subunit of Escherichia coli acetyl-CoA carboxylase.
Biochemistry, 36, 1997
|
|
