6Z2F
 
 | |
5XAV
 
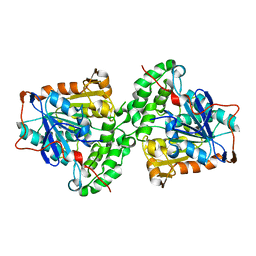 | | Structure of PhaC from Chromobacterium sp. USM2 | | Descriptor: | Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Arsad, H, Samian, M.R, Sudesh, K, Hakoshima, T. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics
Sci Rep, 7, 2017
|
|
6KRJ
 
 | |
6KSR
 
 | |
5XUX
 
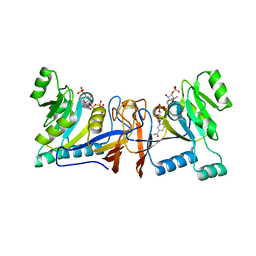 | | Crystal structure of Rib7 from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7A5C
 
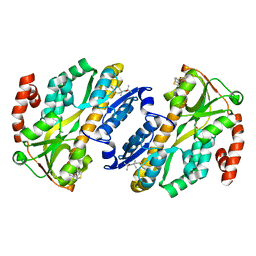 | |
7A5Q
 
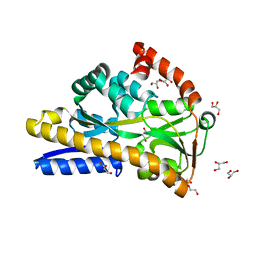 | | Crystal structure of VcSiaP bound to sialic acid | | Descriptor: | DctP family TRAP transporter solute-binding subunit, GLYCEROL, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schneberger, N, Peter, M.F, Hagelueken, G. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Triggering Closure of a Sialic Acid TRAP Transporter Substrate Binding Protein through Binding of Natural or Artificial Substrates.
J.Mol.Biol., 433, 2020
|
|
5XV5
 
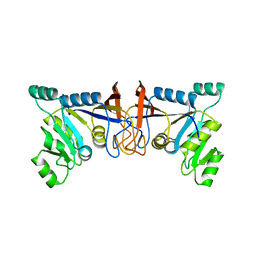 | | Crystal structure of Rib7 mutant S88E from Methanosarcina mazei | | Descriptor: | Conserved protein | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
5XV0
 
 | | Crystal structure of Rib7 mutant D33N from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6VFD
 
 | | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan synthase mutant Q114A in complex with cesium ion at the metal coordination site and 2-aminophenol quinonoid at the enzyme beta site.
To be Published
|
|
6YUD
 
 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
5XV2
 
 | | Crystal structure of Rib7 mutant D33A from Methanosarcina mazei | | Descriptor: | Conserved protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yeh, T.M, Chen, S.C, Chang, T.H, Huang, M.F, Liaw, S.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7ALN
 
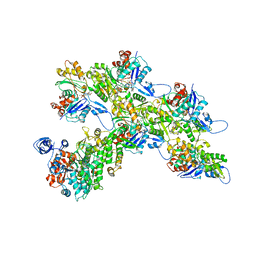 | | Cryo-EM structure of the divergent actomyosin complex from Plasmodium falciparum Myosin A in the Rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Robert-Paganin, J, Xu, X.-P, Swift, M.F, Auguin, D, Robblee, J.P, Lu, H, Fagnant, P.M, Krementsova, E.B, Trybus, K.M, Houdusse, A, Volkmann, N, Hanein, D. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity.
Nat Commun, 12, 2021
|
|
6K3C
 
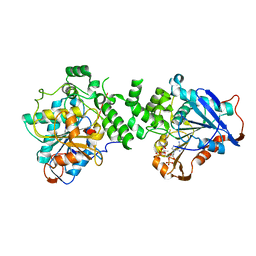 | | Crystal structure of class I PHA synthase (PhaC) mutant from Chromobacterium sp. USM2 bound to Coenzyme A. | | Descriptor: | COENZYME A, Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Hakoshima, T. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.074 Å) | | Cite: | Asymmetric Open-Closed Dimer Mechanism of Polyhydroxyalkanoate Synthase PhaC.
Iscience, 23, 2020
|
|
6W3R
 
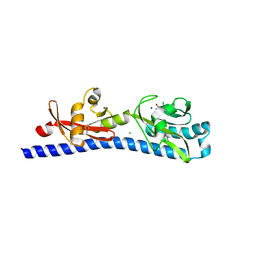 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with 3-methylisoleucine | | Descriptor: | 3-methyl-L-alloisoleucine, CHLORIDE ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6VKO
 
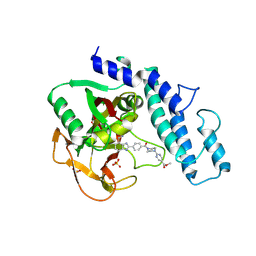 | |
6VNT
 
 | | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution.
To be Published
|
|
6W3Y
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-alanine | | Descriptor: | ALANINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
5L2H
 
 | | Crystal Structure of W26A mutant of anti-EGFR Centyrin P54AR4-83v2 | | Descriptor: | Centyrin, GLYCEROL | | Authors: | Cardoso, R.M.F, Goldberg, S.D, O Neil, K.T, Gilliland, G.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8013 Å) | | Cite: | Engineering a targeted delivery platform using Centyrins.
Protein Eng. Des. Sel., 29, 2016
|
|
5LFR
 
 | |
5LFV
 
 | |
5LJ3
 
 | | Structure of the core of the yeast spliceosome immediately after branching | | Descriptor: | CEF1, CLF1, CWC15, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
5LF5
 
 | |
7UVR
 
 | | Crystal structure of human ClpP protease in complex with TR-65 | | Descriptor: | 3-{[(10R)-4-[(4-chlorophenyl)methyl]-5-oxo-1,2,4,5,8,9-hexahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-7(6H)-yl]methyl}benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVU
 
 | | Crystal structure of human ClpP protease in complex with TR-107 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
