6NB1
 
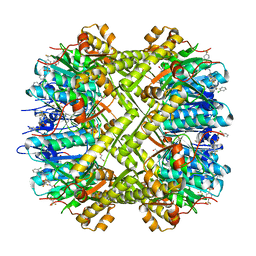 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | Authors: | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NAY
 
 | |
5HBZ
 
 | |
5HF5
 
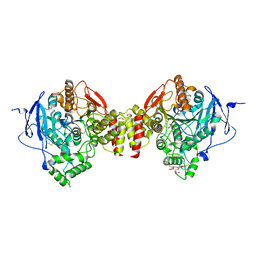 | | Crystal structure of human acetylcholinesterase in complex with paraoxon in the unaged state (predominant acyl loop conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
6NAW
 
 | |
5HFA
 
 | | Crystal structure of human acetylcholinesterase in complex with paraoxon and 2-PAM | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
6NRH
 
 | | Crystal Structure of human PARP-1 ART domain bound inhibitor UTT63 | | Descriptor: | 3-hydroxy-2-({4-[4-(pyrimidin-2-yl)piperazine-1-carbonyl]phenyl}methyl)-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
6NPK
 
 | | Structure of the TM domain | | Descriptor: | Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2019-08-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6NRJ
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT93 | | Descriptor: | (2Z)-2-[(4-{[2-(1H-benzimidazol-2-yl)ethyl]carbamoyl}phenyl)methylidene]-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
8IWA
 
 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWC
 
 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
6NE0
 
 | | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | | Descriptor: | CRISPR RNA (60-MER), CRISPR target DNA (44-MER), CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chowdhury, S, Rollins, M.F, Carter, J, Golden, S.M, Miettinen, H.M, Santiago-Frangos, A, Faith, D, Lawrence, M.C, Wiedenheft, B, Lander, G.C. | | Deposit date: | 2018-12-15 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry.
Mol. Cell, 74, 2019
|
|
6NTU
 
 | |
6NPJ
 
 | | Structure of the NKCC1 CTD | | Descriptor: | Sodium-potassium-chloride cotransporter 1 | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
6O1H
 
 | | Tryptophan synthase Q114A mutant in complex with N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the enzyme alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid at the enzyme beta site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and [3-hydroxy-2-methyl-5-phosphonooxymethyl-pyridin-4-ylmethyl]-serine (PLS) at the beta-site.
To be Published
|
|
6NAQ
 
 | | Crystal structure of Neisseria meningitidis ClpP protease in Apo form | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, POTASSIUM ION | | Authors: | Houry, W.A, Mabanglo, M.F, Pai, E.F, Eger, B.T, Bryson, S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6NRG
 
 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT57 | | Descriptor: | 2-{[3-fluoro-4-(1H-tetrazol-5-yl)phenyl]methyl}-3-hydroxy-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
5IOJ
 
 | |
6NPH
 
 | | Structure of NKCC1 TM domain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Feng, L, Liao, M.F, Orlando, B, Zhang, J.R. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-31 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the cation-chloride cotransporter NKCC1.
Nature, 572, 2019
|
|
3C2G
 
 | | Crystal complex of SYS-1/POP-1 at 2.5A resolution | | Descriptor: | Pop-1 8-residue peptide, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-24 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
8OHN
 
 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
5IHZ
 
 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
5IJK
 
 | | Crystal structure of anti-gliadin 1002-1E03 Fab fragment in complex of peptide PLQPEQPFP | | Descriptor: | 1E03 Fab fragment heavy chain, 1E03 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
6OFQ
 
 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori in complex with peptide STSA | | Descriptor: | Heme-binding protein A / AI-2 binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-03-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
