4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1R0Y
 
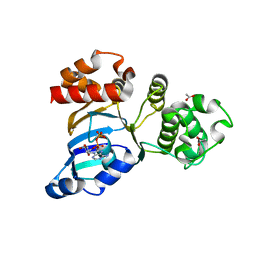 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
6ZQZ
 
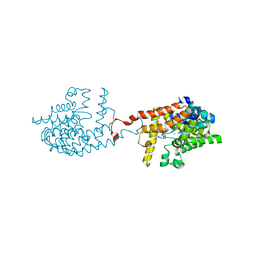 | | [1,2,4]Triazolo[1,5-a]pyrimidine Phosphodiesterase 2 Inhibitors | | Descriptor: | 5-[bis(fluoranyl)methyl]-7-[(3~{S})-1-[(2-chloranyl-6-methyl-pyridin-4-yl)methyl]piperidin-3-yl]-[1,2,4]triazolo[1,5-a]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Tresadern, G, Leonard, P.M. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | [1,2,4]Triazolo[1,5- a ]pyrimidine Phosphodiesterase 2A Inhibitors: Structure and Free-Energy Perturbation-Guided Exploration.
J.Med.Chem., 63, 2020
|
|
1R0X
 
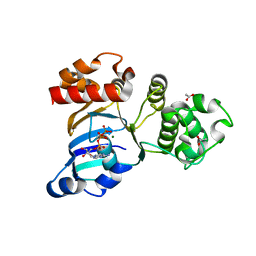 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
1R0W
 
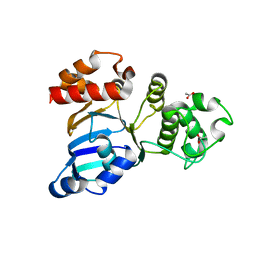 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) apo | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
3KJQ
 
 | | Caspase 8 with covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,8R)-2-(3-carboxypropyl)-8-(2-{[(4-chlorophenyl)acetyl]amino}ethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
1QOG
 
 | | FERREDOXIN MUTATION S47A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1QOF
 
 | | FERREDOXIN MUTATION Q70K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
8YBF
 
 | | Crystal structure of canine distemper virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | Authors: | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | Deposit date: | 2024-02-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
3ZUM
 
 | | Photosynthetic Reaction Centre Mutant with Phe L146 replaced with Ala | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
2AMX
 
 | | Crystal structure of Plasmodium Yoelii Adenosine deaminase (PY02076) | | Descriptor: | COBALT (II) ION, UNKNOWN ATOM OR ION, adenosine deaminase | | Authors: | Dong, A, Vedadi, M, Wasney, G, Zhao, Y, Lew, J, Alam, Z, Melone, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-10 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3ZUW
 
 | | Photosynthetic Reaction Centre Mutant with TYR L128 replaced with HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
1R4C
 
 | |
8LDH
 
 | |
3BE2
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzamide inhibitor | | Descriptor: | N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Gu, Y, Rose, P, Zhao, H. | | Deposit date: | 2007-11-16 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Naphthamides as novel and potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: design, synthesis, and evaluation.
J.Med.Chem., 51, 2008
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2AFN
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE AND A COPPER SITE MUTANT, M150E, THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
6ZJ3
 
 | | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis | | Descriptor: | 18S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Matzov, D, Halfon, H, Zimmerman, E, Rozenberg, H, Bashan, A, Gray, M.W, Yonath, A.E, Shalev-Benami, M. | | Deposit date: | 2020-06-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis.
Nucleic Acids Res., 48, 2020
|
|
3K22
 
 | | Glucocorticoid Receptor with Bound alaninamide 10 with TIF2 peptide | | Descriptor: | Glucocorticoid receptor, N-[(1R)-2-amino-1-methyl-2-oxoethyl]-3-(6-methyl-4-{[3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propyl]amino}-1H-indazol-1-yl)benzamide, Transcriptional Intermediary Factor 2, ... | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V0I
 
 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
4W9C
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(oxazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 2) | | Descriptor: | (4R)-1-(3,3-dimethylbutanoyl)-4-hydroxy-N-[4-(1,3-oxazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Hewitt, S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
4M65
 
 | | In situ thermolysin crystallized on a MiTeGen micromesh with asparagine ligand | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ
co-crystallization of proteins plus fragment libraries on
pin-mounted data-collection micromeshes
Acta Crystallogr.,Sect.D, D70
|
|
4URH
 
 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
3ZYH
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6ZK6
 
 | | Protein Phosphatase 1 (PP1) T320E mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Chembiochem, 22, 2021
|
|
