4AM3
 
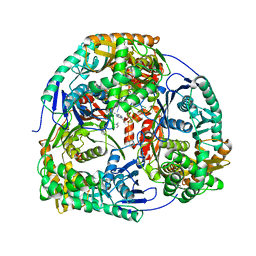 | | Crystal structure of C. crescentus PNPase bound to RNA | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-03-07 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
3GCM
 
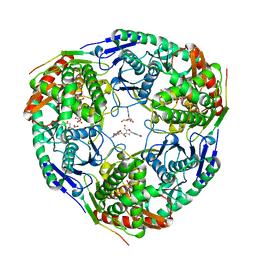 | |
4AID
 
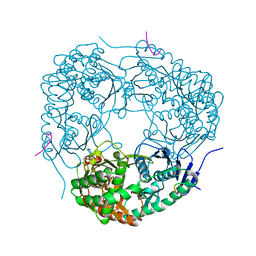 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
8CIB
 
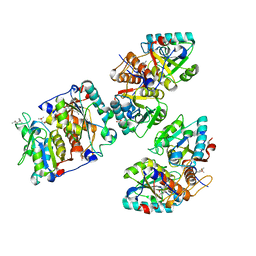 | | Structural and functional analysis of the Pseudomonas aeruginosa PA1677 protein | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Cysteine hydrolase, ... | | 著者 | Sonnleitner, E, Brear, P, Luisi, B.F, Blasi, U. | | 登録日 | 2023-02-09 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Catabolite repression control protein antagonist, a novel player in Pseudomonas aeruginosa carbon catabolite repression control.
Front Microbiol, 14, 2023
|
|
1W08
 
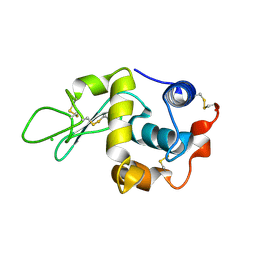 | | STRUCTURE OF T70N HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME | | 著者 | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | 登録日 | 2004-06-02 | | 公開日 | 2004-06-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
4OXP
 
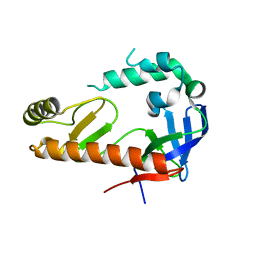 | |
4AIM
 
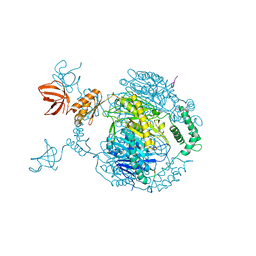 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | 分子名称: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | 著者 | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | 登録日 | 2012-02-10 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
7P06
 
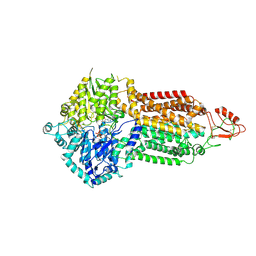 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | 著者 | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | 登録日 | 2021-06-29 | | 公開日 | 2021-11-10 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | 著者 | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | 登録日 | 2021-06-29 | | 公開日 | 2021-11-10 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | 著者 | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | 登録日 | 2021-06-29 | | 公開日 | 2021-11-10 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
6O1L
 
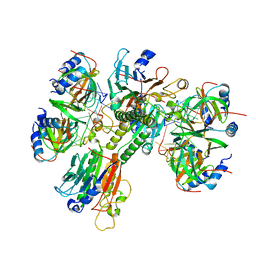 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression Hfq-Crc-amiE 2:3:2 complex | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
6O1K
 
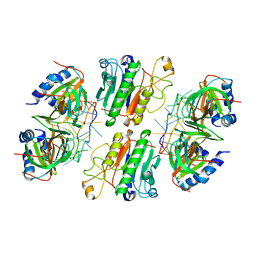 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:2:2 complex (core complex) | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression
Elife, 8, 2019
|
|
5NC5
 
 | |
6O1M
 
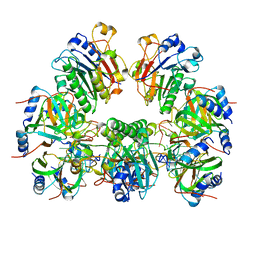 | | Architectural principles for Hfq/Crc-mediated regulation of gene expression. Hfq-Crc-amiE 2:4:2 complex | | 分子名称: | Catabolite repression control protein, RNA (5'-R(*AP*AP*AP*AP*AP*UP*AP*AP*CP*AP*AP*CP*AP*AP*GP*AP*GP*G)-3'), RNA-binding protein Hfq | | 著者 | Pei, X.Y, Dendooven, T, Sonnleitner, E, Chen, S, Blasi, U, Luisi, B.F. | | 登録日 | 2019-02-20 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Architectural principles for Hfq/Crc-mediated regulation of gene expression.
Elife, 8, 2019
|
|
5V5S
 
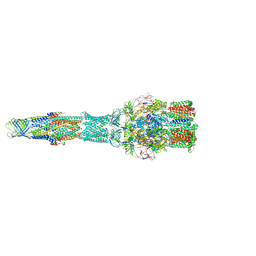 | | multi-drug efflux; membrane transport; RND superfamily; Drug resistance | | 分子名称: | Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, Outer membrane protein TolC | | 著者 | wang, Z, fan, G, Hryc, C.F, Blaza, J.N, Serysheva, I.I, Schmid, M.F, Chiu, W, Luisi, B.F, Du, D. | | 登録日 | 2017-03-15 | | 公開日 | 2017-04-19 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | An allosteric transport mechanism for the AcrAB-TolC Multidrug Efflux Pump.
Elife, 6, 2017
|
|
6TPQ
 
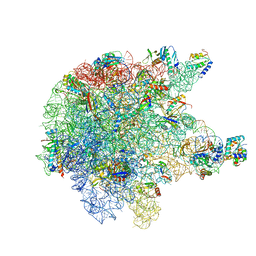 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
6TNN
 
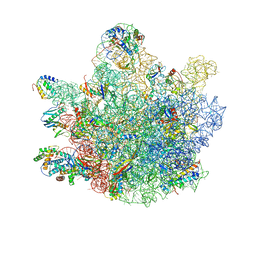 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | 分子名称: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | 登録日 | 2019-12-09 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
4ATO
 
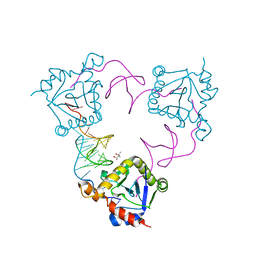 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | 著者 | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | 登録日 | 2012-05-09 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3H8A
 
 | |
297D
 
 | |
285D
 
 | |
2YJN
 
 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | 分子名称: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | 著者 | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.091 Å) | | 主引用文献 | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
4O7J
 
 | |
7P03
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | 分子名称: | Pleiotropic ABC efflux transporter of multiple drugs | | 著者 | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | 登録日 | 2021-06-29 | | 公開日 | 2021-11-10 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
4OWG
 
 | |
