8F40
 
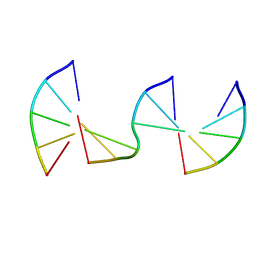 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EPF
 
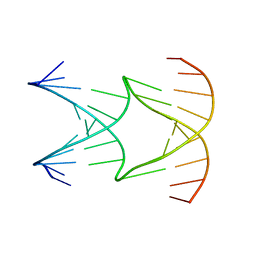 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*GP*GP*TP*GP*GP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*AP*GP*CP*CP*GP*AP*AP*CP*CP*TP*G)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8EP8
 
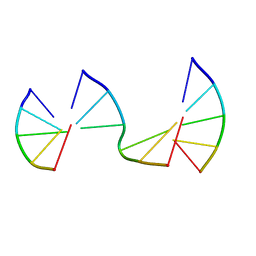 | | Engineering Crystals with Tunable Symmetries from 14- or 16-Base-Long DNA Strands | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*GP*C)-3') | | Authors: | Zhang, C, Zhao, J, Lu, B, Sha, R, Seeman, N.C, Noinaj, N, Mao, C. | | Deposit date: | 2022-10-05 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering DNA Crystals toward Studying DNA-Guest Molecule Interactions.
J.Am.Chem.Soc., 145, 2023
|
|
8G4G
 
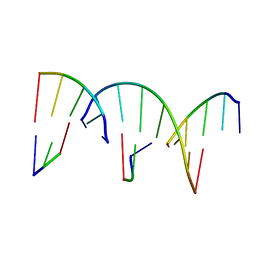 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
5VKZ
 
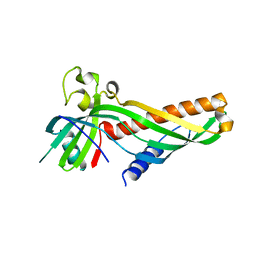 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
7U3R
 
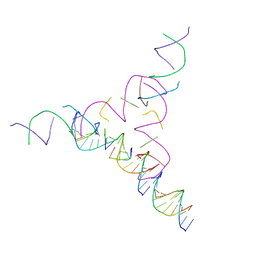 | | [2T7+10] Self-assembling tensegrity triangle with two turns of DNA and the sticky end attachment of a one-turn linker per axis, with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*TP*GP*CP*TP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7U3Q
 
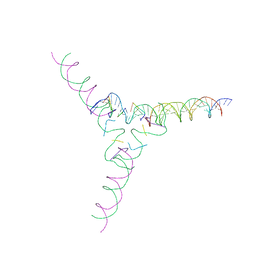 | | [4T7] Self-assembling tensegrity triangle with four turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (29-MER), DNA (42-MER), DNA (5'-D(P*AP*CP*A)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P, Huang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.32 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
6BGG
 
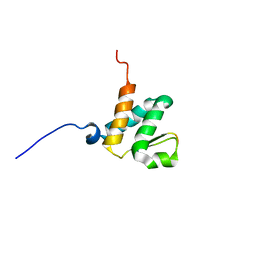 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | Descriptor: | Bromodomain-containing protein 3, CHD4 | | Authors: | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | Deposit date: | 2017-10-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
7U3P
 
 | | [3T7] Self-assembling tensegrity triangle with three turns of DNA per axis with R3 symmetry | | Descriptor: | DNA (31-MER), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Woloszyn, K, Vecchioni, S, Lu, B, Ma, Y, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.06 Å) | | Cite: | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
7F7E
 
 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
7F3Q
 
 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
7F7H
 
 | | SARS-CoV-2 S protein RBD in complex with A8-1 Fab | | Descriptor: | Heavy chain of A8-1 Fab, Light chain of A8-1 Fab, Spike glycoprotein S1 | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | High throughput isolation of potent neutralizing antibodies from convalescent COVID-19 patients.
To Be Published
|
|
