3G9W
 
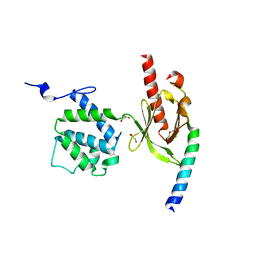 | | Crystal Structure of Talin2 F2-F3 in Complex with the Integrin Beta1D Cytoplasmic Tail | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Integrin beta-1D, ... | | Authors: | Anthis, N.J, Wegener, K.L, Ye, F, Kim, C, Lowe, E.D, Vakonakis, I, Bate, N, Critchley, D.R, Ginsberg, M.H, Campbell, I.D. | | Deposit date: | 2009-02-15 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | The structure of an integrin/talin complex reveals the basis of inside-out signal transduction
Embo J., 28, 2009
|
|
6NLR
 
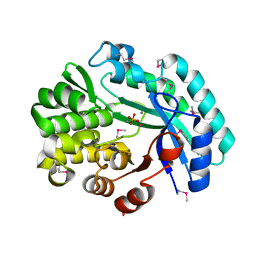 | | Crystal structure of the putative histidinol phosphatase hisK from Listeria monocytogenes with trinuclear metals determined by PIXE revealing sulphate ion in active site. Based on PIXE analysis and original date from 3DCP | | Descriptor: | CALCIUM ION, COBALT (II) ION, FE (III) ION, ... | | Authors: | Snell, E.H, Garman, E.F, Lowe, E.D. | | Deposit date: | 2019-01-09 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput PIXE as an Essential Quantitative Assay for Accurate Metalloprotein Structural Analysis: Development and Application.
J.Am.Chem.Soc., 142, 2020
|
|
6OE2
 
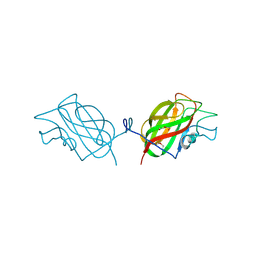 | |
6OBY
 
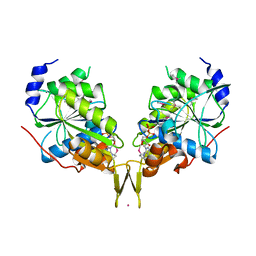 | |
5JZB
 
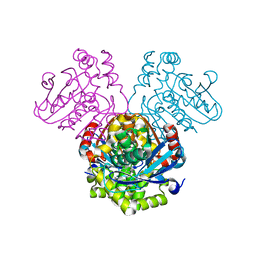 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZ9
 
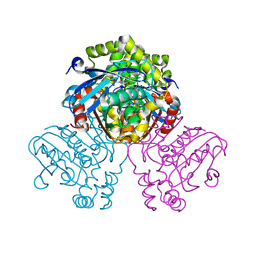 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
4X0L
 
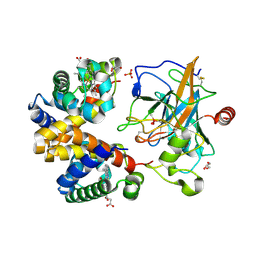 | | Human haptoglobin-haemoglobin complex | | Descriptor: | CACODYLATE ION, GLYCEROL, Haptoglobin, ... | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
4V3D
 
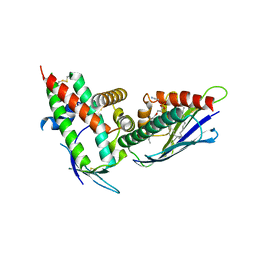 | | The CIDRa domain from HB3var03 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
4UTF
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-isofagomine and alpha- 1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, ... | | Authors: | Cuskin, F, Lowe, E.C, Temple, M.J, Zhu, Y, Pudlo, N.A, Cameron, E.A, Urs, K, Thompson, A.J, Cartmell, A, Rogowski, A, Tolbert, T, Piens, K, Bracke, D, Vervecken, W, Hakki, Z, Speciale, G, Munoz-Munoz, J.L, Pena, M.J, McLean, R, Suits, M.D, Boraston, A.B, Atherly, T, Ziemer, C.J, Williams, S.J, Davies, G.J, Abbott, D.W, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
4X0J
 
 | | Trypanosoma brucei haptoglobin-haemoglobin receptor | | Descriptor: | Haptoglobin-hemoglobin receptor | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
3LT5
 
 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
3LTW
 
 | | The structure of mycobacterium marinum arylamine n-acetyltransferase in complex with hydralazine | | Descriptor: | 1-hydrazinophthalazine, Arylamine N-acetyltransferase Nat, FORMIC ACID | | Authors: | Abuhammad, A.M, Lowe, E.D, Fullam, E, Noble, M, Garman, E.F, Sim, E. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site
Protein Cell, 1, 2010
|
|
3R6W
 
 | | paAzoR1 binding to nitrofurazone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Ryan, A, Kaplan, K, Laurieri, N, Lowe, E, Sim, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Activation of nitrofurazone by azoreductases: multiple activities in one enzyme.
Sci Rep, 1, 2011
|
|
3BLH
 
 | | Crystal Structure of Human CDK9/cyclinT1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Baumli, S, Lolli, G, Lowe, E.D, Johnson, L.N. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation
Embo J., 27, 2008
|
|
3BLR
 
 | | Crystal Structure of Human CDK9/cyclinT1 in complex with Flavopiridol | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, Cell division protein kinase 9, Cyclin-T1, ... | | Authors: | Baumli, S, Lolli, G, Lowe, E.D, Johnson, L.N. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation
Embo J., 27, 2008
|
|
6XZ1
 
 | | Conjugate of the HECT domain of HUWE1 with ubiquitin | | Descriptor: | HECT, UBA and WWE domain containing 1, isoform CRA_a, ... | | Authors: | Liu, B, Seenivasan, A, Nair, R, Chen, D, Lowe, E.D, Lorenz, S. | | Deposit date: | 2020-01-31 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Acs Chem.Biol., 16, 2021
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
3EJH
 
 | | Crystal Structure of the Fibronectin 8-9FnI Domain Pair in Complex with a Type-I Collagen Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen type-I a1 chain, Fibronectin, ... | | Authors: | Erat, M.C, Lowe, E.D, Campbell, I.D, Vakonakis, I. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and structural analysis of type I collagen sites in complex with fibronectin fragments.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4N65
 
 | | Crystal structure of paAzoR1 bound to anthraquinone-2-sulphonate | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Kaplan, E, Crescente, V, Lowe, E, Preston, G.M, Sim, E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Identification of NAD(P)H Quinone Oxidoreductase Activity in Azoreductases from P. aeruginosa: Azoreductases and NAD(P)H Quinone Oxidoreductases Belong to the Same FMN-Dependent Superfamily of Enzymes.
Plos One, 9, 2014
|
|
8AH3
 
 | |
