2XQB
 
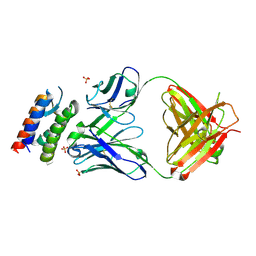 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
3SNF
 
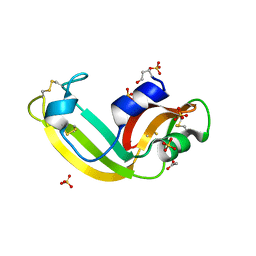 | | Onconase, atomic resolution crystal structure | | Descriptor: | ACETATE ION, Protein P-30, SULFATE ION | | Authors: | Holloway, D.E, Singh, U.P, Shogen, K, Acharya, K.R. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of Onconase at 1.1 angstrom resolution--insights into substrate binding and collective motion.
Febs J., 278, 2011
|
|
1XIA
 
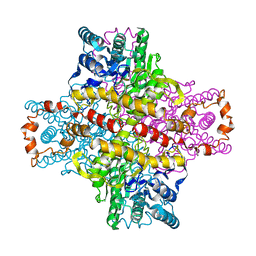 | |
1UN5
 
 | | ARH-II, AN ANGIOGENIN/RNASE A CHIMERA | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2004-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Studies on Structural Features that Determine the Enzymatic Specificity and Potency of Human Angiogenin: Thr44, Thr80 and Residues 38-41
Biochemistry, 43, 2004
|
|
1UN3
 
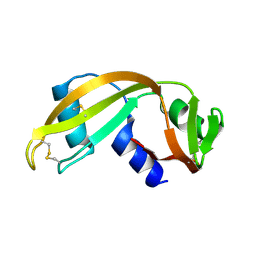 | |
1UN4
 
 | |
1GV7
 
 | | ARH-I, an angiogenin/RNase A chimera | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Shapiro, R, Hares, M.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Guest-Host Crosstalk in an Angiogenin-RNase A Chimeric Protein
Biochemistry, 41, 2002
|
|
2BWK
 
 | | Murine angiogenin, sulphate complex | | Descriptor: | ANGIOGENIN, SULFATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BWL
 
 | | Murine angiogenin, phosphate complex | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4UV4
 
 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
4G6V
 
 | | CdiA-CT/CdiI toxin and immunity complex from Burkholderia pseudomallei | | Descriptor: | Adhesin/hemolysin, BROMIDE ION, CdiI | | Authors: | Morse, R.P, Nikolakakis, K, Willet, J, Gerrick, E, Low, D.A, Hayes, C.S, Goulding, C.W. | | Deposit date: | 2012-07-19 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G6U
 
 | | CdiA-CT/CdiI toxin and immunity complex from Escherichia coli | | Descriptor: | ACETATE ION, CHLORIDE ION, EC869 CdiA-CT, ... | | Authors: | Morse, R.P, Nikolakakis, K, Willet, J, Gerrick, E, Low, D.A, Hayes, C.S, Goulding, C.W. | | Deposit date: | 2012-07-19 | | Release date: | 2012-12-12 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TS1
 
 | |
4A7J
 
 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | Descriptor: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | Authors: | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | Deposit date: | 2011-11-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
4Q7O
 
 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | Descriptor: | BROMIDE ION, FORMIC ACID, Immunity protein | | Authors: | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
8GI6
 
 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
2XFX
 
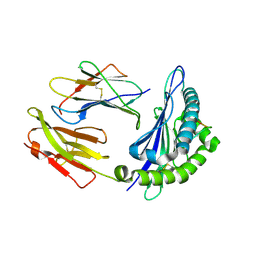 | | cattle MHC class I N01301 presenting an 11mer from Theileria parva | | Descriptor: | BETA-2-MICROGLOBULIN, MHC CLASS 1, UNCHARACTERIZED PROTEIN | | Authors: | Macdonald, I.K, Harkiolaki, M, Hunt, L, Morrison, W.I, Connelley, T, Graham, S.P, Jones, E.Y, Flower, D.R, Ellis, S.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mhc Class I Bound to an Immunodominant Theileria Parva Epitope Demonstrates Unconventional Presentation to T Cell Receptors.
Plos Pathog., 6, 2010
|
|
6ZPM
 
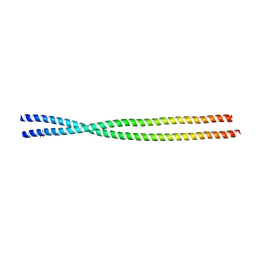 | | Crystal structure of the unconventional kinetochore protein Trypanosoma cruzi KKT4 coiled coil domain | | Descriptor: | THREONINE, Trypanosoma cruzi KKT4 117-218 | | Authors: | Ludzia, P, Lowe, D.E, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
4TS1
 
 | |
5A4C
 
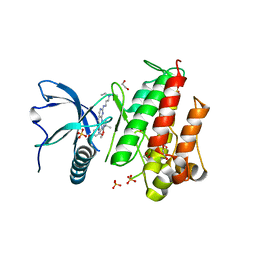 | | FGFR1 ligand complex | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
5A46
 
 | | FGFR1 in complex with dovitinib | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
8G5A
 
 | |
