1A0C
 
 | | XYLOSE ISOMERASE FROM THERMOANAEROBACTERIUM THERMOSULFURIGENES | | Descriptor: | COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | Deposit date: | 1997-11-28 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
8GI6
 
 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
8G5B
 
 | |
8G5A
 
 | |
4XIA
 
 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
5FZT
 
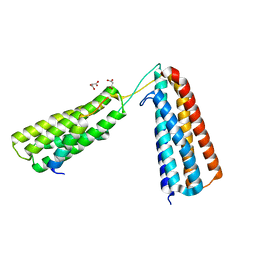 | | The crystal structure of R7R8 in complex with a DLC1 fragment. | | Descriptor: | MALONATE ION, RHO GTPASE-ACTIVATING PROTEIN 7, TALIN-1 | | Authors: | Zacharchenko, T, Qian, X, Goult, B.T, Jethwa, D, Almeida, T, Ballestrem, C, Critchley, D.R, Lowy, D.R, Barsukov, I.L. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ld Motif Recognition by Talin: Structure of the Talin-Dlc1 Complex.
Structure, 24, 2016
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
3J6R
 
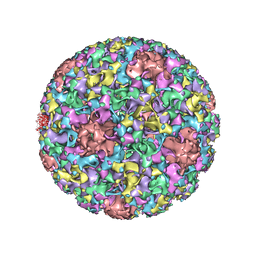 | | Electron cryo-microscopy of Human Papillomavirus Type 16 capsid | | Descriptor: | Major capsid protein L1 | | Authors: | Cardone, G, Moyer, A.L, Cheng, N, Thompson, C.D, Dvoretzky, I, Lowy, D.R, Schiller, J.T, Steven, A.C, Buck, C.B, Trus, B.L. | | Deposit date: | 2014-03-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Maturation of the human papillomavirus 16 capsid.
MBio, 5, 2014
|
|
4OLR
 
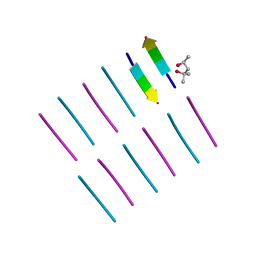 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
4ONK
 
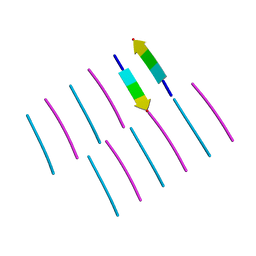 | | [Leu-5]-Enkephalin mutant - YVVFL | | Descriptor: | [Leu-5]-Enkephalin mutant - YVVFL | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
5XIA
 
 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
1H5N
 
 | | DMSO REDUCTASE MODIFIED BY THE PRESENCE OF DMS AND AIR | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM(VI) ION, ... | | Authors: | Bailey, S, Adams, B, Smith, A.T, Richards, R.L, Lowe, D.J, Bray, R.C. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactions of Dimethyl Sulphoxide Reductase in the Presence of Dimethylsulphide and the Structure of the Dimethylsulphide-Modified Enzyme
Biochemistry, 40, 2001
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
3TS1
 
 | |
4CHA
 
 | |
4DAJ
 
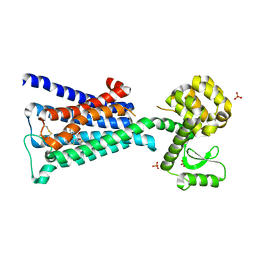 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
2XFX
 
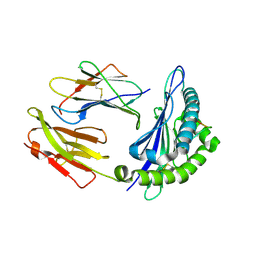 | | cattle MHC class I N01301 presenting an 11mer from Theileria parva | | Descriptor: | BETA-2-MICROGLOBULIN, MHC CLASS 1, UNCHARACTERIZED PROTEIN | | Authors: | Macdonald, I.K, Harkiolaki, M, Hunt, L, Morrison, W.I, Connelley, T, Graham, S.P, Jones, E.Y, Flower, D.R, Ellis, S.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mhc Class I Bound to an Immunodominant Theileria Parva Epitope Demonstrates Unconventional Presentation to T Cell Receptors.
Plos Pathog., 6, 2010
|
|
3ZBW
 
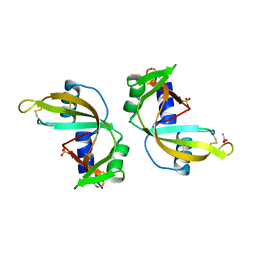 | | Crystal Structure of murine Angiogenin-3 | | Descriptor: | ACETIC ACID, ANGIOGENIN-3, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
3ZBV
 
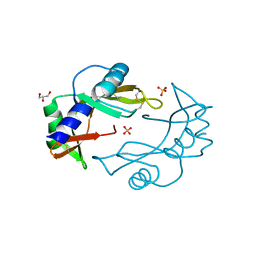 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
1TIE
 
 | |
1T15
 
 | | Crystal Structure of the Brca1 BRCT Domains in Complex with the Phosphorylated Interacting Region from Bach1 Helicase | | Descriptor: | BRCA1 interacting protein C-terminal helicase 1, Breast cancer type 1 susceptibility protein | | Authors: | Clapperton, J.A, Manke, I.A, Lowery, D.M, Ho, T, Haire, L.F, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2004-04-15 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XLI
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | 5-thio-alpha-D-glucopyranose, D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLD
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
