5TG1
 
 | | 1.40 A resolution structure of Norovirus 3CL protease in complex with the a m-chlorophenyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17S)-17-(3-chlorophenyl)-7-(hydroxymethyl)-4-(2-methylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
5TG2
 
 | | 1.75 A resolution structure of Norovirus 3CL protease in complex with the a n-pentyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17R)-7-(hydroxymethyl)-4-(2-methylpropyl)-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
7K7E
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 7.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7K7D
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 6.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
5WEJ
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a dipeptidyl oxazolidinone-based inhibitor | | Descriptor: | (2S)-2-{(5S)-5-[(3-chlorophenyl)methyl]-2-oxo-1,3-oxazolidin-3-yl}-4-methyl-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}pentanamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Rathnayake, A.D, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design, synthesis and evaluation of oxazolidinone-based inhibitors of norovirus 3CL protease.
Eur J Med Chem, 143, 2017
|
|
7K7B
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7KNF
 
 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7K7C
 
 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.5 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
4TOF
 
 | | 1.65A resolution structure of BfrB (C89S, K96C) crystal form 1 from Pseudomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterioferritin, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4LQT
 
 | | 1.10A resolution crystal structure of a superfolder green fluorescent protein (W57A) mutant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
4LQU
 
 | | 1.60A resolution crystal structure of a superfolder green fluorescent protein (W57G) mutant | | Descriptor: | Green fluorescent protein | | Authors: | Lovell, S, Xia, Y, Vo, B, Battaile, K.P, Egan, C, Karanicolas, J. | | Deposit date: | 2013-07-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The designability of protein switches by chemical rescue of structure: mechanisms of inactivation and reactivation.
J.Am.Chem.Soc., 135, 2013
|
|
7K5H
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-66 | | Descriptor: | 4-{[3-(3-chloro-5-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K5E
 
 | | 1.75 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor JAG-5-7 | | Descriptor: | 4-{[(5-fluoro-2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K5F
 
 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-50 | | Descriptor: | 4-{[(3-chloro-5-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7K5G
 
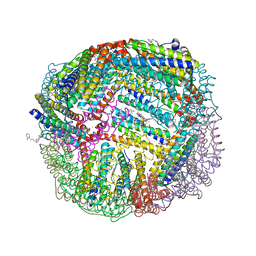 | | 1.95 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-28 | | Descriptor: | 4-{[3-(2-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
4O6T
 
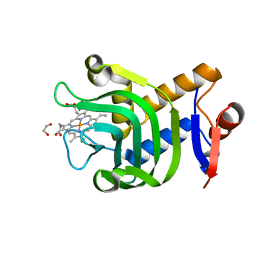 | | 1.25A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, pH 5.4) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
3RIX
 
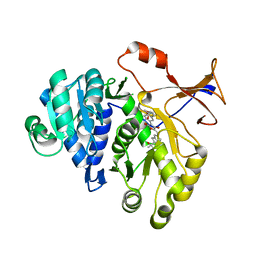 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
4O6S
 
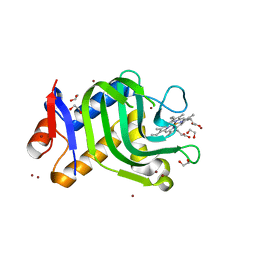 | | 1.32A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, Zinc Bound) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6Q
 
 | | 0.95A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (Y75A mutant) | | Descriptor: | FORMIC ACID, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4O6U
 
 | | 0.89A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
3R9I
 
 | | 2.6A resolution structure of MinD complexed with MinE (12-31) peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
8TGB
 
 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
3R9J
 
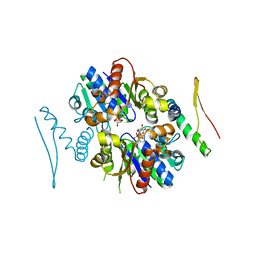 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3PZF
 
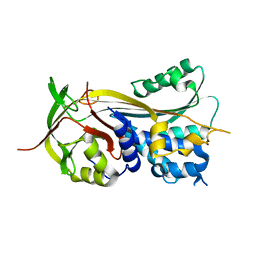 | | 1.75A resolution structure of Serpin-2 from Anopheles gambiae | | Descriptor: | Serpin 2 | | Authors: | Lovell, S, Battaile, K.P, An, C, Michel, K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of native Anopheles gambiae serpin-2, a negative regulator of melanization in mosquitoes.
Proteins, 79, 2011
|
|
