7TQ2
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ3
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9BK3
 
 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
7RT0
 
 | |
8W1E
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, SULFATE ION | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1F
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer, Mg bound) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4B
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4A
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 11c | | Descriptor: | (1R,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(7-cyano-7-azaspiro[3.5]nonan-2-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Z
 
 | | Structure of MERS 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T48
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 9c | | Descriptor: | (1R,2S)-2-{[N-({[(2r,4R)-7-acetyl-7-azaspiro[3.5]non-5-en-2-yl]oxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[7-(phenylacetyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7TQ5
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 10d | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[(1R,2R)-2-(4-methoxyphenyl)cyclopropyl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
9BK2
 
 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
3RIX
 
 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
7K5G
 
 | | 1.95 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-28 | | Descriptor: | 4-{[3-(2-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7KNG
 
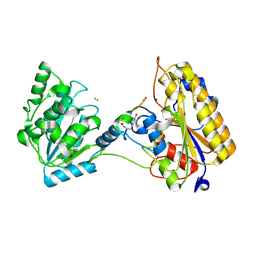 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7K5H
 
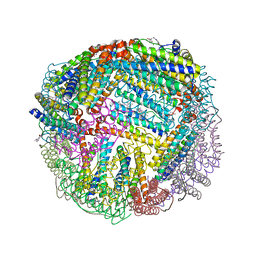 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-66 | | Descriptor: | 4-{[3-(3-chloro-5-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K5E
 
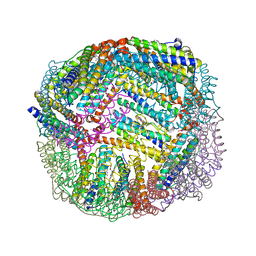 | | 1.75 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor JAG-5-7 | | Descriptor: | 4-{[(5-fluoro-2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K5F
 
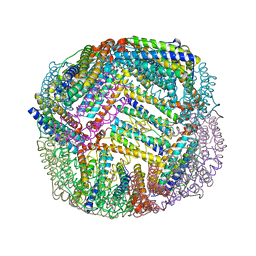 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-50 | | Descriptor: | 4-{[(3-chloro-5-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7K7C
 
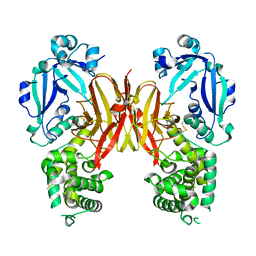 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.5 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
7KNF
 
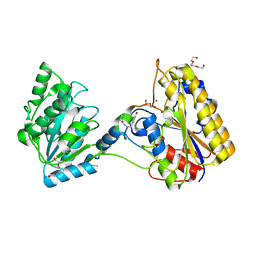 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7K7B
 
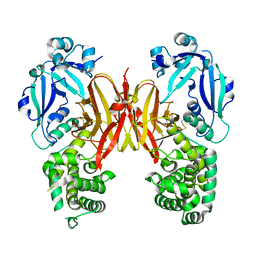 | | Crystal structure of diphtheria toxin from crystals obtained at pH 5.0 | | Descriptor: | Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rodnin, M.V, Ladokhin, A.S. | | Deposit date: | 2020-09-22 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Toxins, 12, 2020
|
|
