1GFM
 
 | | OMPF PORIN (MUTANT D113G) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFP
 
 | | OMPF PORIN (MUTANT R42C) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFN
 
 | | OMPF PORIN DELETION (MUTANT DELTA 109-114) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFO
 
 | | OMPF PORIN (MUTANT R132P) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
1GFQ
 
 | | OMPF PORIN (MUTANT R82C) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MATRIX PORIN OUTER MEMBRANE PROTEIN F | | Authors: | Lou, K.-L, Schirmer, T. | | Deposit date: | 1996-05-08 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis.
J.Biol.Chem., 271, 1996
|
|
5JI0
 
 | | PPARgamma-RXRalpha(S427F) heterodimer in complex with SRC-1, rosiglitazone, and 9-cis-retanoic acid | | Descriptor: | (9cis)-retinoic acid, 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), ... | | Authors: | Bloudoff, K, Larsen, N.A. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PPARgamma-RXRalpha(S427F) heterodimer in complex with SRC-1, rosiglitazone, and 9-cis-retanoic acid
To Be Published
|
|
5T3E
 
 | |
5DUA
 
 | |
5DU9
 
 | | First condensation domain of the calcium-dependent antibiotic synthetase in complex with substrate analogue 2a | | Descriptor: | (2S)-2-amino-N-butyl-propanamide, CDA peptide synthetase I, CHLORIDE ION, ... | | Authors: | Bloudoff, K, Alonzo, D.A, Schmeing, T.M. | | Deposit date: | 2015-09-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical Probes Allow Structural Insight into the Condensation Reaction of Nonribosomal Peptide Synthetases.
Cell Chem Biol, 23, 2016
|
|
1V1H
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a short linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
4JN3
 
 | |
1V1I
 
 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a long linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
4JN5
 
 | |
6P8H
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P21 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8G
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8E
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1, ... | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6P8F
 
 | | Crystal structure of CDK4 in complex with CyclinD1 and P27 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1B, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
3FEC
 
 | | Crystal structure of human Glutamate Carboxypeptidase III (GCPIII/NAALADase II), pseudo-unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Barinka, C, Lubkowski, J, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FEE
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
3FED
 
 | | The high resolution structure of human glutamate carboxypeptidase III (GCPIII/NAALADase II) in complex with a transition state analog of Glu-Glu | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-amino-3-carboxypropyl](hydroxy)phosphoryl]methyl}pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubkowski, J, Barinka, C, Hlouchova, K. | | Deposit date: | 2008-11-28 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insight into the evolutionary and pharmacologic homology of glutamate carboxypeptidases II and III
Febs J., 276, 2009
|
|
7LY6
 
 | | Structure of a trans-acting NRPS oxidase, BmdC, involved in bacillamide biosynthesis | | Descriptor: | BmdC, NRPS oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fortinez, C.M, Bloudoff, K, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
6DN2
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1354 SPLIT RNA | | Descriptor: | 4-{benzyl[2-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)ethyl]amino}butanoic acid, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN3
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN1
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6N8G
 
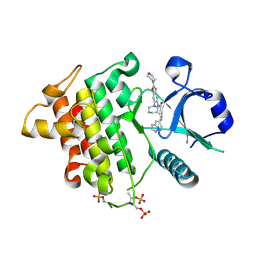 | | IRAK4 bound to benzoxazole compound | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[2-(morpholin-4-yl)-6-(piperidin-1-yl)-1,3-benzoxazol-5-yl]-6-(1H-pyrrolo[2,3-b]pyridin-5-yl)pyridine-2-carboxamide | | Authors: | Larsen, N.A, Bloudoff, K, Subramanian, V, Dobrzanska, M, Gluza, K. | | Deposit date: | 2018-11-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | To be published
To Be Published
|
|
