1VDY
 
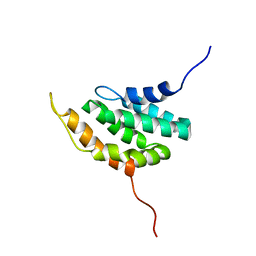 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
2DCQ
 
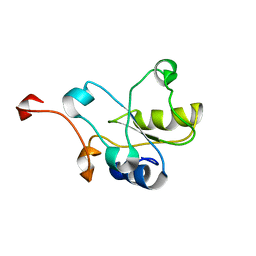 | |
2DCP
 
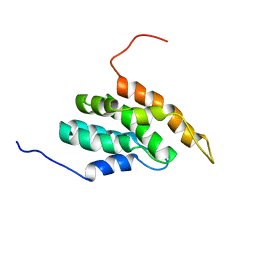 | |
2DCR
 
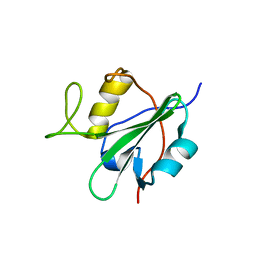 | |
2H9X
 
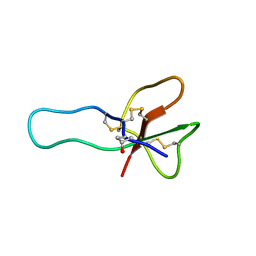 | | NMR structure for the CgNa toxin from the sea anemone Condylactis gigantea | | Descriptor: | Toxin CgNa | | Authors: | Lopez-Mendez, B, Perez-Castells, J, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2006-06-12 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | CgNa, a type I toxin from the giant Caribbean sea anemone Condylactis gigantea shows structural similarities to both type I and II toxins, as well as distinctive structural and functional properties(1).
Biochem.J., 406, 2007
|
|
2KAC
 
 | |
2MQK
 
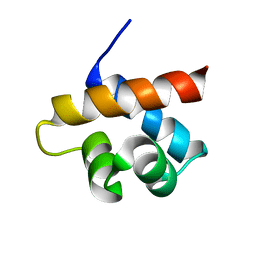 | |
1VEE
 
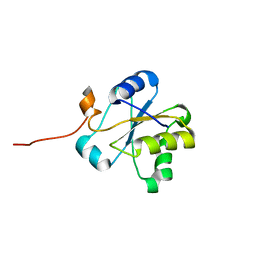 | | NMR structure of the hypothetical rhodanese domain At4g01050 from Arabidopsis thaliana | | Descriptor: | proline-rich protein family | | Authors: | Pantoja-Uceda, D, Lopez-Mendez, B, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-01-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the rhodanese homology domain At4g01050(175-295) from Arabidopsis thaliana
Protein Sci., 14, 2005
|
|
7PQ3
 
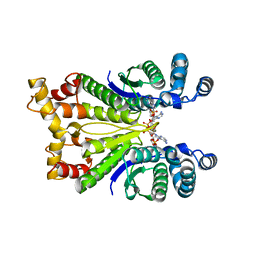 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
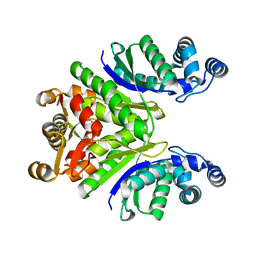 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ6
 
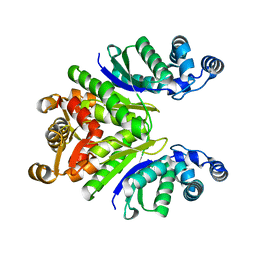 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQA
 
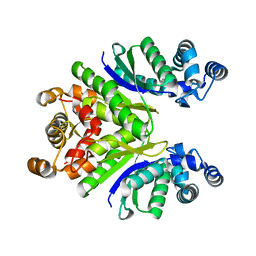 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7Z55
 
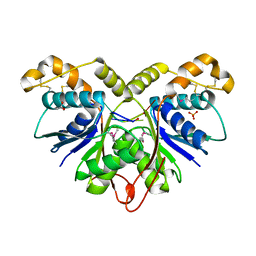 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in complex with its substrate | | Descriptor: | ACETATE ION, CRISPR-associated protein, Cyclic RNA cA4, ... | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
7Z56
 
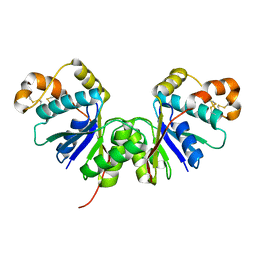 | | Crystal Structure of the Ring Nuclease 0455 from Sulfolobus islandicus (Sis0455) in its apo form | | Descriptor: | CRISPR-associated protein | | Authors: | Molina, R, Martin-Garcia, R, Lopez-Mendez, B, Jensen, A.L.G, Marchena-Hurtado, J, Stella, S, Montoya, G. | | Deposit date: | 2022-03-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.
Nucleic Acids Res., 50, 2022
|
|
8AA5
 
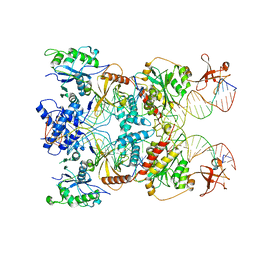 | | Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon) | | Descriptor: | LE_PolyA, LE_Target, RE_PolyA, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Lopez-Mendez, B, Stutzke, L.S, Fuglsang, A, Stella, S, Montoya, G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon.
Nat Commun, 13, 2022
|
|
4CJ9
 
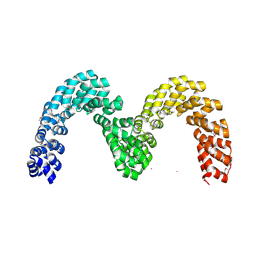 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in its apo form | | Descriptor: | BURRH, SELENIUM ATOM | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2GLH
 
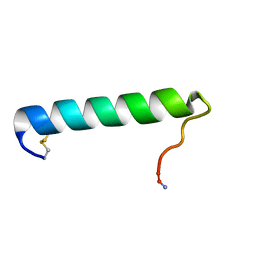 | | Solution Conformation of Salmon Calcitonin in Sodium Dodecyl Sulfate Micelles | | Descriptor: | Calcitonin-1 | | Authors: | Andreotti, G, Lopez-Mendez, B, Amodeo, P, Morelli, M.A, Nakamuta, H, Motta, A. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of salmon calcitonin bioactivity: the role of the Leu-based amphipathic alpha-helix.
J.Biol.Chem., 281, 2006
|
|
2GLG
 
 | | NMR structure of the [L23,A24]-sCT mutant | | Descriptor: | Calcitonin-1 | | Authors: | Andreotti, G, Lopez-Mendez, B, Amodeo, P, Morelli, M.A, Nakamuta, H, Motta, A. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-20 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of salmon calcitonin bioactivity: the role of the Leu-based amphipathic alpha-helix.
J.Biol.Chem., 281, 2006
|
|
4CJA
 
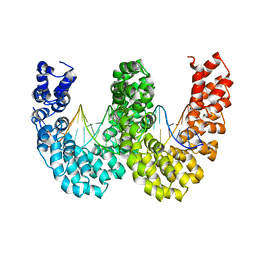 | | BurrH DNA-binding protein from Burkholderia rhizoxinica in complex with its target DNA | | Descriptor: | 5'-D(*DTP*AP*TP*AP*AP*CP*GP*TP*AP*TP*TP*TP*GP*CP *TP*TP*CP*TP*CP*TP*TP*AP*AP)-3', 5'-D(*DTP*TP*AP*AP*GP*AP*GP*AP*AP*GP*CP*AP*AP*DP *TP*AP*CP*GP*TP*TP*AP*TP*AP)-3', BURRH | | Authors: | Stella, S, Molina, R, Lopez-Mendez, B, Campos-Olivas, R, Duchateau, P, Montoya, G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Bud, a Helix-Loop-Helix DNA-Binding Domain for Genome Modification
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
8AXB
 
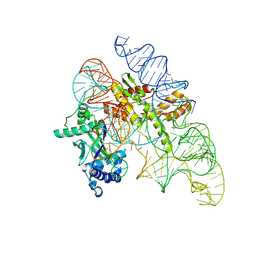 | | Cryo-EM structure of Cas12k-sgRNA binary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, sgRNA | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Molina, R, Pape, T, Lopez-Mendez, B, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
