4QKS
 
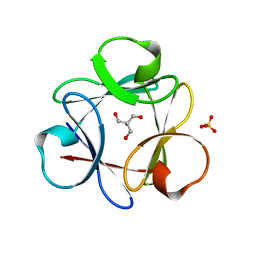 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | 著者 | Longo, L.M, Tenorio, C.A, Blaber, M. | | 登録日 | 2014-06-09 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
4QKR
 
 | |
6LFG
 
 | |
6M6P
 
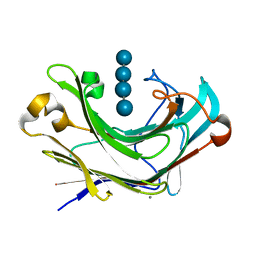 | | Structure of Marine bacterial laminarinase mutant E135A in complex with 1,3-beta-cellotriosyl-glucose | | 分子名称: | CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | 著者 | Yang, J, Xu, Y, Tanokura, M, Long, L, Miyakawa, T. | | 登録日 | 2020-03-16 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6L6Z
 
 | |
6JH5
 
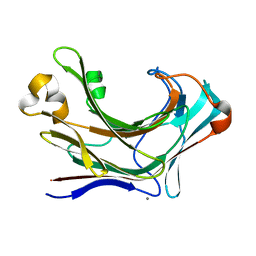 | | Structure of Marine bacterial laminarinase | | 分子名称: | CALCIUM ION, LamCAT | | 著者 | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | 登録日 | 2019-02-17 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | 分子名称: | CALCIUM ION, LamCAT | | 著者 | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | 登録日 | 2019-02-18 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JIA
 
 | | Marine bacterial laminarinase mutant E135A complex with laminaritetraose | | 分子名称: | CALCIUM ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | 著者 | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | 登録日 | 2019-02-20 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
3SNV
 
 | |
6ITC
 
 | | Structure of a substrate engaged SecA-SecY protein translocation machine | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Ma, C.Y, Wu, X.F, Sun, D.J, Park, E.Y, Rapoport, T.A, Gao, N, Long, L. | | 登録日 | 2018-11-21 | | 公開日 | 2019-06-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Structure of the substrate-engaged SecA-SecY protein translocation machine.
Nat Commun, 10, 2019
|
|
8CYI
 
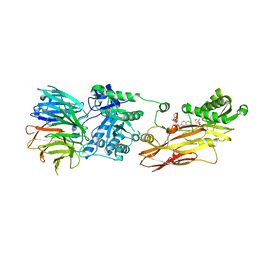 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | 分子名称: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | 著者 | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | 登録日 | 2022-05-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
8QDH
 
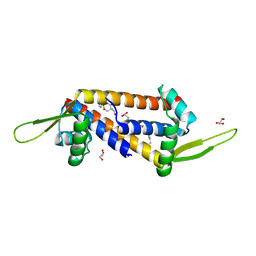 | | Engineered LmrR carrying a cyclic boronate ester formed between Tris and p-boronophenylalanine at position 89 | | 分子名称: | GLYCEROL, Transcriptional regulator, PadR-like family | | 著者 | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | 登録日 | 2023-08-29 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
8QDF
 
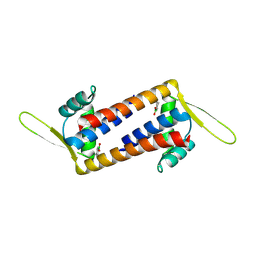 | | Engineered LmrR with Met-89 replaced by para-boronophenylalanine | | 分子名称: | Transcriptional regulator, PadR-like family | | 著者 | Thunnissen, A.M.W.H, Rozeboom, H.J, Longwitz, L, Leveson-Gower, R.B, Roelfes, G. | | 登録日 | 2023-08-29 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Boron catalysis in a designer enzyme.
Nature, 629, 2024
|
|
4OW4
 
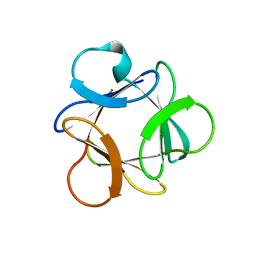 | |
4D8H
 
 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | 著者 | Blaber, M, Longo, L. | | 登録日 | 2012-01-10 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
