6AE9
 
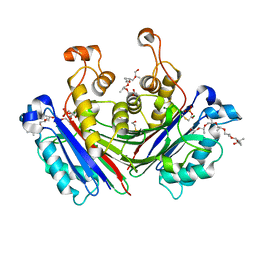 | | X-ray structure of the photosystem II phosphatase PBCP | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Liu, X.Y, Chai, J.C, Ou, X.M, Liu, Z.F. | | Deposit date: | 2018-08-03 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into Substrate Selectivity, Catalytic Mechanism, and Redox Regulation of Rice Photosystem II Core Phosphatase.
Mol Plant, 12, 2019
|
|
7EE6
 
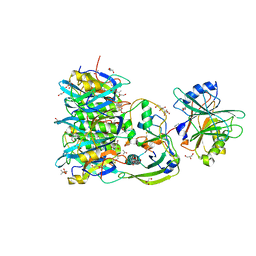 | | Crystal structure of PltC toxin | | Descriptor: | ACETONE, CITRATE ANION, Cytolethal distending toxin subunit B family protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE3
 
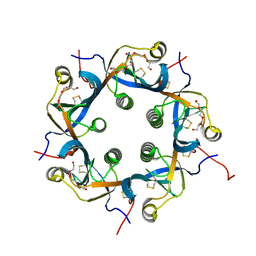 | | Crystal structure of PltC | | Descriptor: | Subtilase cytotoxin subunit B-like protein, TETRAETHYLENE GLYCOL | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE5
 
 | | Crystal structure of Neu5Gc bound PltC | | Descriptor: | N-glycolyl-alpha-neuraminic acid, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, Subtilase cytotoxin subunit B-like protein, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7EE4
 
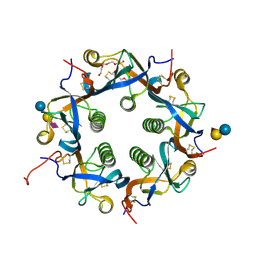 | | Crystal structure of Neu5Ac bound PltC | | Descriptor: | N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Liu, X.Y, Chen, Z, Gao, X. | | Deposit date: | 2021-03-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin.
Mbio, 13, 2022
|
|
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7CYE
 
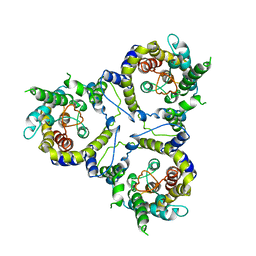 | | Cryo-EM structure of sodium-dependent bicarbonate transporter SbtA from Synechocystis sp. PCC 6803 | | Descriptor: | Slr1512 protein | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
5WTR
 
 | | Crystal structure of a prokaryotic TRIC channel in 0.5 M KCl | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, TRISTEAROYLGLYCEROL, ... | | Authors: | Ou, X.M, Wang, L.F, Yang, H.T, Liu, X.Y, Liu, Z.F. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ion and water binding sites inside an occluded hourglass pore of a TRIC channel
BMC BIOL., 2017
|
|
3JCU
 
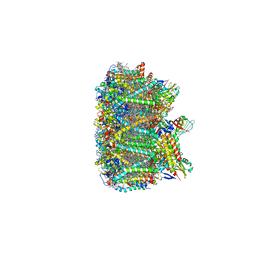 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
5WFU
 
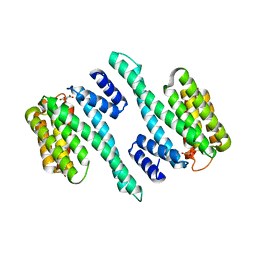 | |
4BYG
 
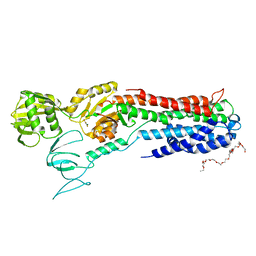 | | ATPase crystal structure | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Pedersen, B.P, Morth, J.P, Wang, J, Gourdon, P, Nissen, P. | | Deposit date: | 2013-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dephosphorylation of Pib-Type Cu(I)-Atpases as Studied by Metallofluoride Complexes
To be Published
|
|
4BEV
 
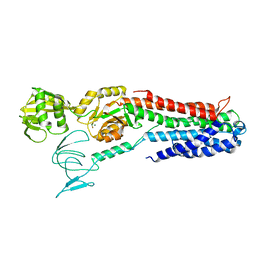 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
6KY5
 
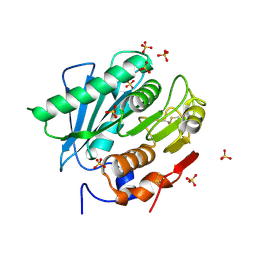 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
4EGS
 
 | |
4F5E
 
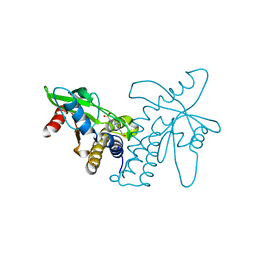 | | Crystal structure of ERIS/STING | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F5D
 
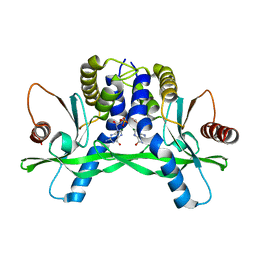 | | ERIS/STING in complex with ligand | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Transmembrane protein 173 | | Authors: | Huang, Y.H, Liu, X.Y, Su, X.D. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for the sensing and binding of cyclic di-GMP by STING
Nat.Struct.Mol.Biol., 19, 2012
|
|
8WYD
 
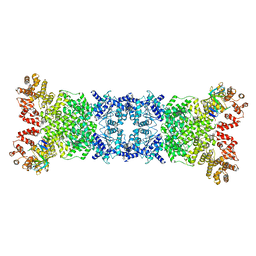 | | Cryo-EM structure of DSR2-DSAD1 complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY9
 
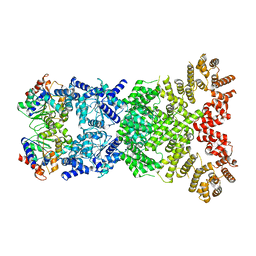 | |
8WYE
 
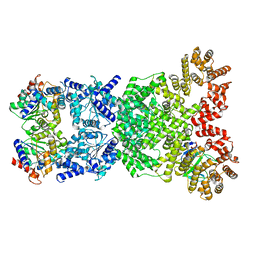 | | Cryo-EM structure of DSR2-DSAD1 (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYA
 
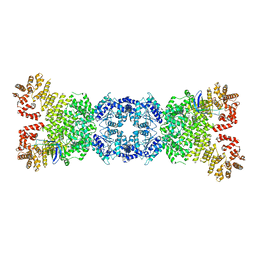 | | Cryo-EM structure of DSR2-tube complex | | Descriptor: | Bacillus phage SPbeta tube protein, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYF
 
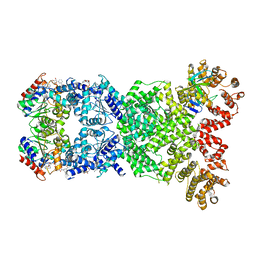 | | Cryo-EM structure of DSR2-DSAD1-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPbeta DSAD1 protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYC
 
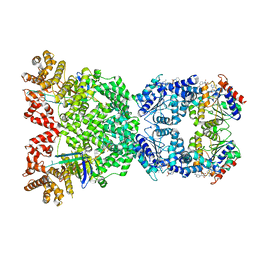 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ (partial) complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYB
 
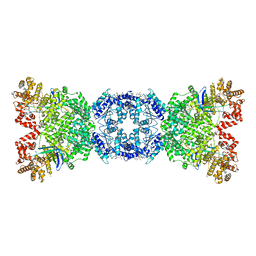 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ complex | | Descriptor: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY8
 
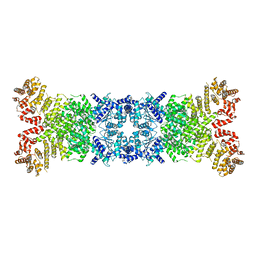 | | Cryo-EM structure of DSR2 apo complex | | Descriptor: | SIR2 family protein | | Authors: | Zhang, J.T, Jia, N, Liu, X.Y. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
4ETI
 
 | |
