7TBL
 
 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
3DBQ
 
 | | Crystal structure of TTK kinase domain | | Descriptor: | Dual specificity protein kinase TTK | | Authors: | Wang, W, Yang, Y.T, Gao, Y.F, Zhu, S.C, Wang, F, Old, W, Xu, Q.B, Resing, K, Ahn, N, Lei, M, Liu, X.D. | | Deposit date: | 2008-06-02 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into Mps1 Kinase Activation
J.CELL.MOL.MED., 13, 2008
|
|
6KY5
 
 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
9EQM
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
5TQR
 
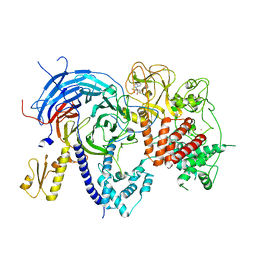 | |
5ZHP
 
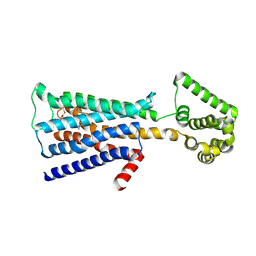 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TBI
 
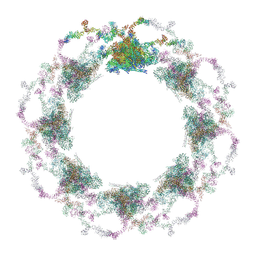 | | Composite structure of the S. cerevisiae nuclear pore complex (NPC) | | Descriptor: | Dyn2, Nic96 R1, Nic96 R2, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5VK3
 
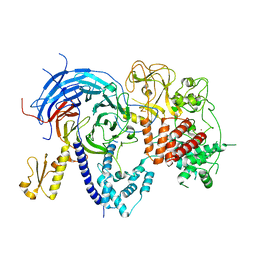 | | Apo ctPRC2 with E840A and K852D mutations in Ezh2 | | Descriptor: | Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12, Polycomb Protein EED, ZINC ION | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-04-20 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
2L8S
 
 | |
5WAK
 
 | | Crystal Structure of a Suz12-Rbbp4 Binary Complex | | Descriptor: | Histone-binding protein RBBP4, Polycomb protein SUZ12, ZINC ION | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
2GYY
 
 | |
7YIK
 
 | |
3CXP
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3CXS
 
 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
4KP3
 
 | | Crystal Structure of MyoVa-GTD in Complex with Two Cargos | | Descriptor: | Melanophilin, RILP-like protein 2, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3PMR
 
 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
7YOQ
 
 | | Recj2 and CMP complex from Methanocaldococcus jannaschii | | Descriptor: | [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)furan-2-yl]methyl dihydrogen phosphate, nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
7YOS
 
 | |
3CXQ
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3R5T
 
 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
6A6O
 
 | | Crystal structure of acetyl ester-xyloside bifunctional hydrolase from Caldicellulosiruptor lactoaceticus | | Descriptor: | Esterase/lipase-like protein | | Authors: | Cao, H, Huang, Y, Sun, L.C, Liu, X, Liu, T.F, Wang, F.Z, Xin, F.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Dual-Substrate Recognition and Catalytic Mechanisms of a Bifunctional Acetyl Ester-Xyloside Hydrolase from Caldicellulosiruptor lactoaceticus.
Acs Catalysis, 9, 2019
|
|
3R5S
 
 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
2GZ1
 
 | | Structure of Aspartate Semialdehyde Dehydrogenase (ASADH) from Streptococcus pneumoniae complexed with NADP | | Descriptor: | Aspartate beta-semialdehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Faehnle, C.R, Le Coq, J, Liu, X, Viola, R.E. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examination of key intermediates in the catalytic cycle of aspartate-beta-semialdehyde dehydrogenase from a gram-positive infectious bacteria.
J.Biol.Chem., 281, 2006
|
|
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
