7LW2
 
 | |
8EOY
 
 | |
7MG0
 
 | |
5HC7
 
 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in complex with S-thiolo-isopentenyldiphosphate | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, prenyltransference for protein | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5HC6
 
 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in apo form | | Descriptor: | SULFATE ION, prenyltransference for protein | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5HC8
 
 | | Crystal structure of lavandulyl diphosphate synthase from Lavandula x intermedia in complex with dimethylallyl diphosphate | | Descriptor: | 2-methylbuta-1,3-diene, 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, MAGNESIUM ION, ... | | Authors: | Liu, M.X, Liu, W.D, Gao, J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure and Function of a "Head-to-Middle" Prenyltransferase: Lavandulyl Diphosphate Synthase
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7RFW
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFR
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFS
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2022-01-05 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
5XO6
 
 | | Crystal structure of a novel ZEN lactonase mutant | | Descriptor: | PENTAETHYLENE GLYCOL, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
2VLC
 
 | | Crystal structure of Natural Cinnamomin (Isoform III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein, beta-D-mannopyranose, ... | | Authors: | Azzi, A, Wang, T, Zhu, D.-W, Zou, Y.-S, Liu, W.-Y, Lin, S.-X. | | Deposit date: | 2008-01-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Native Cinnamomin Isoform III and its Comparison with Other Ribosome Inactivating Proteins.
Proteins: Struct., Funct., Bioinf., 74, 2009
|
|
8V4U
 
 | |
2P4Q
 
 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7SDC
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MI-09 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SDA
 
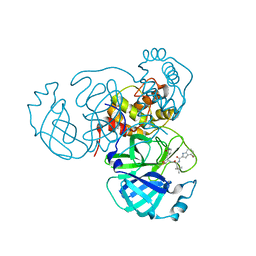 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI49 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SD9
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI48 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4,4-dimethyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-09-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SH8
 
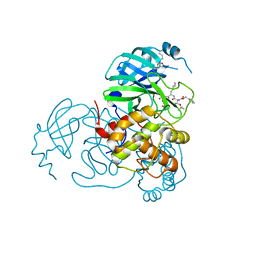 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI88 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3R)-1-{[(2S)-1-(2-acetyl-2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}hydrazinyl)-3-cyclohexyl-1-oxopropan-2-yl]amino}-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-10-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
7SH9
 
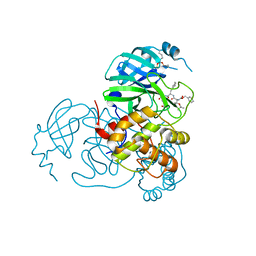 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI86 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3R)-1-({(2S)-1-[2-acetyl-2-(3-amino-3-oxopropyl)hydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2021-10-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the SARS-CoV-2 main protease in complex with inhibitors
To Be Published
|
|
7TE0
 
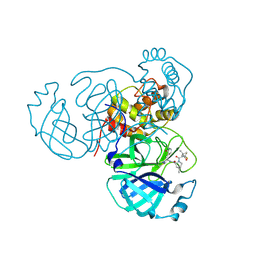 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary and Structural Insights about Potential SARS-CoV-2 Evasion of Nirmatrelvir.
J.Med.Chem., 65, 2022
|
|
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | Descriptor: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSJ
 
 | | Crystal structure of 1,6-APD (2-ANIMOPHENOL-1,6-DIOXYGENASE) complexed with intermediate products | | Descriptor: | (2Z,4Z)-2-imino-6-oxohex-4-enoic acid, (3E)-3-iminooxepin-2(3H)-one, 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, ... | | Authors: | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | Deposit date: | 2012-04-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5Z7J
 
 | | Crystal structure of a lactonase double mutant in complex with ligand l | | Descriptor: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, DI(HYDROXYETHYL)ETHER, Lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
