5NBT
 
 | | Apo structure of p60N/p80C katanin | | 分子名称: | Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | 著者 | Jiang, K, Rezabkova, L, Hua, S, Liu, Q, Capitani, G, Altelaar, A.F.M, Heck, A.J.R, Kammerer, R.A, Steinmetz, M.O, Akhmanova, A. | | 登録日 | 2017-03-02 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
1RSG
 
 | |
1VB0
 
 | | Atomic resolution structure of atratoxin-b, one short-chain neurotoxin from Naja atra | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cobrotoxin b, SULFATE ION | | 著者 | Lou, X, Liu, Q, Teng, M, Niu, L, Huang, Q, Hao, Q. | | 登録日 | 2004-02-20 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
1QWO
 
 | | Crystal structure of a phosphorylated phytase from Aspergillus fumigatus, revealing the structural basis for its heat resilience and catalytic pathway | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, phytase | | 著者 | Xiang, T, Liu, Q, Deacon, A.M, Koshy, M, Kriksunov, I.A, Lei, X.G, Hao, Q, Thiel, D.J. | | 登録日 | 2003-09-03 | | 公開日 | 2004-06-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of a Heat-resilient Phytase from Aspergillus fumigatus, Carrying a Phosphorylated Histidine
J.Mol.Biol., 339, 2004
|
|
1V6P
 
 | | Crystal structure of Cobrotoxin | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Cobrotoxin, ... | | 著者 | Lou, X, Tu, X, Wang, J, Teng, M, Niu, L, Liu, Q, Huang, Q, Hao, Q. | | 登録日 | 2003-12-03 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (0.87 Å) | | 主引用文献 | The atomic resolution crystal structure of atratoxin determined by single wavelength anomalous diffraction phasing
J.Biol.Chem., 279, 2004
|
|
3LQQ
 
 | | Structure of the CED-4 Apoptosome | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Qi, S, Pang, Y, Shi, Y, Yan, N, Liu, Q. | | 登録日 | 2010-02-09 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.534 Å) | | 主引用文献 | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
1TW9
 
 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | 分子名称: | glutathione S-transferase 2 | | 著者 | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | 登録日 | 2004-06-30 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
1XPQ
 
 | | Crystal structure of fms1, a polyamine oxidase from yeast | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, SPERMINE | | 著者 | Huang, Q, Liu, Q, Hao, Q. | | 登録日 | 2004-10-09 | | 公開日 | 2005-04-26 | | 最終更新日 | 2021-12-08 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structures of Fms1 and its complex with spermine reveal substrate specificity.
J.Mol.Biol., 348, 2005
|
|
1YY5
 
 | |
2QKW
 
 | | Structural basis for activation of plant immunity by bacterial effector protein AvrPto | | 分子名称: | Avirulence protein, Protein kinase | | 著者 | Xing, W.M, Zou, Y, Liu, Q, Hao, Q, Zhou, J.M, Chai, J.J. | | 登録日 | 2007-07-11 | | 公開日 | 2007-08-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The structural basis for activation of plant immunity by bacterial effector protein AvrPto
Nature, 449, 2007
|
|
6PBR
 
 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | 分子名称: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | 著者 | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | 登録日 | 2019-06-14 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1ZL9
 
 | | Crystal Structure of a major nematode C.elegans specific GST (CE01613) | | 分子名称: | GLUTATHIONE, glutathione S-transferase 5 | | 著者 | Kriksunov, I.A, Liu, Q, Schuller, D.J, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | 登録日 | 2005-05-05 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structure of a major nematode C.elegans specific GST (CE01613)
To be Published
|
|
2A5Y
 
 | | Structure of a CED-4/CED-9 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Apoptosis regulator ced-9, MAGNESIUM ION, ... | | 著者 | Yan, N, Liu, Q, Hao, Q, Gu, L, Shi, Y. | | 登録日 | 2005-07-01 | | 公開日 | 2005-10-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans.
Nature, 437, 2005
|
|
8IGL
 
 | |
4ED5
 
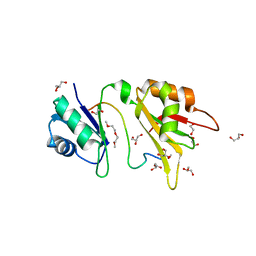 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | 著者 | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | 登録日 | 2012-03-27 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6NQ8
 
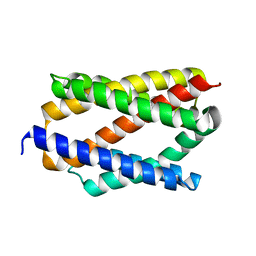 | |
6NQ9
 
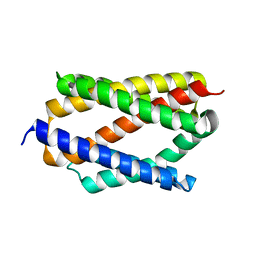 | |
6NQ7
 
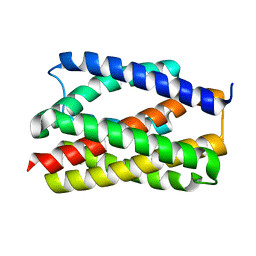 | |
3I9O
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ribo-2'F-ADP ribose | | 分子名称: | ADP-ribosyl cyclase, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4S)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | 著者 | Graeff, R, Liu, Q, Kriksunov, I.A, Kotaka, M, Oppenheimer, N, Hao, Q, Lee, H.C. | | 登録日 | 2009-07-12 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mechanism of cyclizing NAD to cyclic ADP-ribose by ADP-ribosyl cyclase and CD38
J.Biol.Chem., 284, 2009
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | 著者 | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | 登録日 | 2022-01-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | 著者 | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | 登録日 | 2013-03-15 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | 著者 | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | 登録日 | 2013-03-14 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JNF
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | 分子名称: | Hsp70 CHAPERONE DnaK | | 著者 | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | 登録日 | 2013-03-15 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.621 Å) | | 主引用文献 | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6OE8
 
 | | The crystal structure of hyper-thermostable AgUricase mutant K12C/E286C | | 分子名称: | MALONATE ION, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Shi, Y, Wang, T, Zhou, X.E, Liu, Q, Jiang, Y, Xu, H.E. | | 登録日 | 2019-03-27 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-based design of a hyperthermostable AgUricase for hyperuricemia and gout therapy.
Acta Pharmacol.Sin., 40, 2019
|
|
1Y00
 
 | | Solution structure of the Carbon Storage Regulator protein CsrA | | 分子名称: | Carbon storage regulator | | 著者 | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2004-11-13 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
