6O3N
 
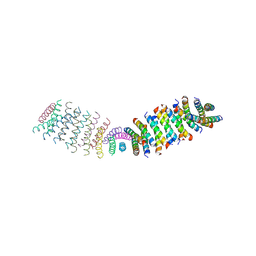 | |
4R99
 
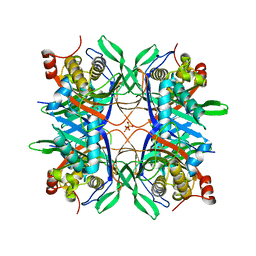 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | SULFATE ION, Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
8IYA
 
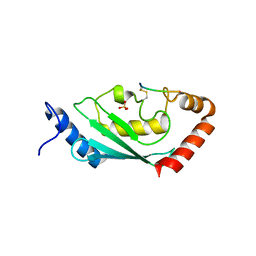 | | Complex of SETDB1-derived peptide bound to UBE2E1 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, SULFATE ION, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Du, Y.X, Liu, L. | | Deposit date: | 2023-04-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1.
Nat Commun, 15, 2024
|
|
8J1R
 
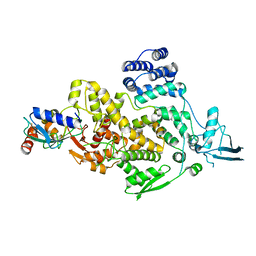 | | cryo-EM structures of Ufd4 in complex with Ubc4-Ub | | Descriptor: | Ubiquitin fusion degradation protein 4, Ubiquitin-conjugating enzyme E2 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Cai, H.Y, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Visualization of HECT-E3 Ufd4 accepting and transferring Ubiquitin to Form K29/K48-branched Polyubiquitination on N-degron. bioRxiv,doi: ttps://doi.org/10.1101/2023.05.23.542033
To Be Published
|
|
8J1P
 
 | | Cryo-EM structure of Ufd4 in complex with K29/48 triUb | | Descriptor: | Ubiquitin, Ubiquitin fusion degradation protein 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural Insights into the Molecular Mechanism of Ufd4-catalyzed Elongation of K48-linked Ubiquitin Chain through Lys29 Linkage
To Be Published
|
|
7XD7
 
 | | The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The pre-Tet-C state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD5
 
 | | The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD6
 
 | | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7O25
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE from T. maritima (reaction triggered in the crystal) | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1P
 
 | | [FeFe]-hydrogenase maturase HydE from T. Maritima (C-ter stretch absent) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O26
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (5'dA + Methionine) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBON MONOXIDE, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1S
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Wild-type protein) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1T
 
 | | Fe(CO)2CNCl species bound [HydE from T. Maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
7O1O
 
 | | Complex-B bound [FeFe]-hydrogenase maturase HydE fromT. Maritima (Auxiliary cluster deleted variant) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, CYANIDE ION, ... | | Authors: | Rohac, R, Martin, L, Liu, L, Basu, D, Tao, L, Britt, R.D, Rauchfuss, T, Nicolet, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the [FeFe]-Hydrogenase Maturase HydE Bound to Complex-B.
J.Am.Chem.Soc., 143, 2021
|
|
3VTR
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 E328A complexed with TMG-chitotriomycin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Chen, W, Liu, L, Shen, X, Yang, Q. | | Deposit date: | 2012-06-02 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into cellulolytic and chitinolytic enzymes revealing crucial residues of insect beta-N-acetyl-D-hexosaminidase
Plos One, 7, 2012
|
|
7X7T
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zheng, Q, Li, S, Zhang, T, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7V
 
 | | Cryo-EM structure of SARS-CoV spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X7U
 
 | | Cryo-EM structure of SARS-CoV-2 Delta variant spike protein in complex with three nAbs X01, X10 and X17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, X01 heavy chain, ... | | Authors: | Sun, H, Liu, L, Zhang, T, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | The neutralizing breadth of antibodies targeting diverse conserved epitopes between SARS-CoV and SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1VPR
 
 | |
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEX
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
8KAG
 
 | | Crystal structure of SpyCas9 in complex with sgRNA and target RNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, RNA (5'-R(*CP*CP*AP*CP*UP*UP*CP*AP*AP*UP*UP*AP*GP*AP*AP*CP*AP*CP*GP*GP*AP*CP*C)-3'), RNA (98-MER) | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
8KAK
 
 | |
