2VF2
 
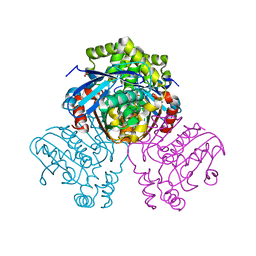 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4ZRD
 
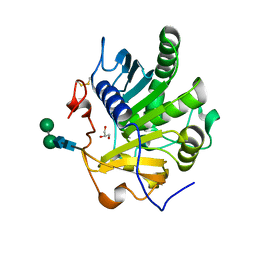 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4YY9
 
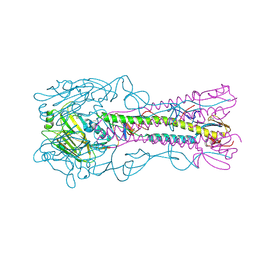 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4YY7
 
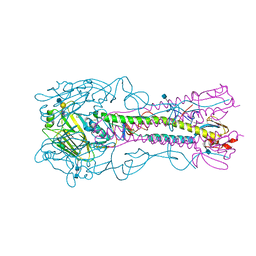 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYA
 
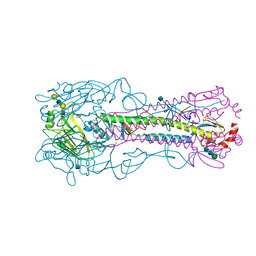 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYB
 
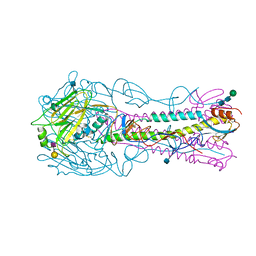 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
5A3I
 
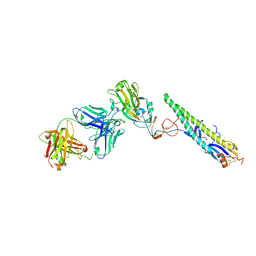 | | Crystal Structure of a Complex formed between FLD194 Fab and Transmissible Mutant H5 Haemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HAEMAGGLUTININ HA1 FAB HEAVY CHAIN, ... | | Authors: | Xiong, X, Corti, D, Liu, J, Pinna, D, Foglierini, M, Calder, L.J, Martin, S.R, Lin, Y.P, Walker, P.A, Collins, P.J, Monne, I, Suguitan Jr, A.L, Santos, C, Temperton, N.J, Subbarao, K, Lanzavecchia, A, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Complexes Formed by H5 Influenza Hemagglutinin with a Potent Broadly Neutralizing Human Monoclonal Antibody.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1XS2
 
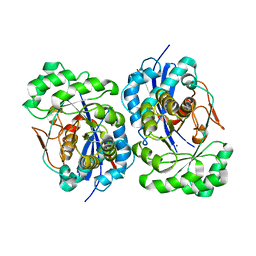 | | Structural Basis for Catalytic Racemization and Substrate Specificity of an N-Acylamino Acid Racemase Homologue from Deinococcus radiodurans | | Descriptor: | MAGNESIUM ION, N-Acylamino Acid Racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
3GE9
 
 | |
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
3AMH
 
 | | crystal structure of cellulase 12A from Thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMN
 
 | | E134C-Cellobiose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMP
 
 | | E134C-Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
1ZG2
 
 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
3AMM
 
 | | Cellotetraose complex of cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Liu, J.-R, Guo, R.-T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate-binding mode of cellulase 12A from Thermotoga maritima
Proteins, 79, 2011
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
1ZVA
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZVB
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV7
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | CHLORIDE ION, spike glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
