8J19
 
 | | Cryo-EM structure of the LY237-bound GPR84 receptor-Gi complex | | Descriptor: | 6-nonylpyridine-2,4-diol, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J1A
 
 | | Cryo-EM structure of the GPR84 receptor-Gi complex with no ligand modeled | | Descriptor: | Antibody fragment ScFv16, G-protein coupled receptor 84, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
8J18
 
 | | Cryo-EM structure of the 3-OH-C12-bound GPR84 receptor-Gi complex | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Antibody fragment ScFv16, G-protein coupled receptor 84, ... | | Authors: | Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Nat Commun, 14, 2023
|
|
2JBM
 
 | | QPRTASE STRUCTURE FROM HUMAN | | Descriptor: | NICOTINATE-NUCLEOTIDE PYROPHOSPHORYLASE, S,R MESO-TARTARIC ACID | | Authors: | Liu, H, Naismith, J.H. | | Deposit date: | 2006-12-08 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Quinolinate Phosphoribosyltransferase (Hqprtase) from Homo Sapiens.
J.Mol.Biol., 373, 2007
|
|
5GSK
 
 | | Crystal structure of duplex DNA3 in complex with Hg(II) and Sr(II) | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION, STRONTIUM ION | | Authors: | Liu, H.H, Wang, R, Yao, Q.Q, Cheng, Y.Q, Yang, C, Luo, Q, Wu, B.X, Li, J.X, Ma, J.B, Sheng, J, Gan, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5XNQ
 
 | | Crystal structures of human SALM5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
8H89
 
 | | Capsid of Ralstonia phage GP4 | | Descriptor: | Major capsid protein, Virion associated protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-10-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Viruses, 14, 2022
|
|
6DKU
 
 | | Crystal structure of Myotis VP35 mutant of interferon inhibitory domain | | Descriptor: | VP35 | | Authors: | Liu, H, Ginell, G.M, Keefe, L.J, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conservation of Structure and Immune Antagonist Functions of Filoviral VP35 Homologs Present in Microbat Genomes.
Cell Rep, 24, 2018
|
|
7EYB
 
 | | core proteins | | Descriptor: | Internal virion protein gp14, Internal virion protein gp15, Peptidoglycan transglycosylase gp16 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY7
 
 | | bacteriophage T7 tail complex | | Descriptor: | Internal virion protein gp14, Tail fiber protein, Tail tubular protein gp11, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY9
 
 | | tail proteins | | Descriptor: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY8
 
 | | portal | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY6
 
 | | The portal protein (GP8) of bacteriophage T7 | | Descriptor: | Portal protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2021-05-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TUK
 
 | |
7TUL
 
 | |
4LFI
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Liu, H. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KTR
 
 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
5XNP
 
 | | Crystal structures of human SALM5 in complex with human PTPdelta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.729 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5XGP
 
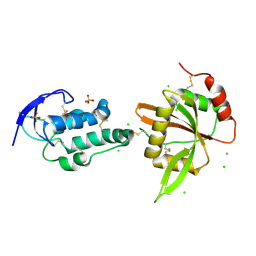 | | structure of Sizzled from Xenopus laevis at 2.08 angstroms resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Secreted Xwnt8 inhibitor sizzled | | Authors: | Liu, H, Li, Z, Xu, F. | | Deposit date: | 2017-04-15 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | The crystal structure of full-length Sizzled from Xenopus laevis yields insights into Wnt-antagonistic function of secreted Frizzled-related proteins
J. Biol. Chem., 292, 2017
|
|
4EXP
 
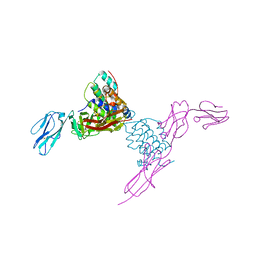 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4EXN
 
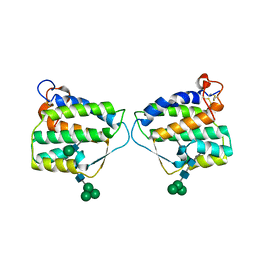 | | Crystal structure of mouse Interleukin-34 | | Descriptor: | Interleukin-34, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4JR7
 
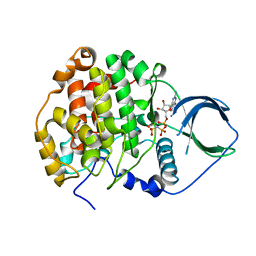 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | Descriptor: | Casein kinase II subunit alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4FI1
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H, Wang, H, Teng, M, Li, X. | | Deposit date: | 2012-06-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of scCK2 alpha in complex with ATP
To be Published
|
|
2O26
 
 | | Structure of a class III RTK signaling assembly | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor, ... | | Authors: | Liu, H, Chen, X, Focia, P.J, He, X. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases.
Embo J., 26, 2007
|
|
