2Q7W
 
 | | Structural Studies Reveals the Inactivation of E. coli L-aspartate aminotransferase (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms at pH 6.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2QB2
 
 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (s)-4,5-dihydro-2thiophenecarboylic acid (SADTA) via two mechanisms (at pH 7.0). | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
8J4Z
 
 | |
8J7O
 
 | |
8J73
 
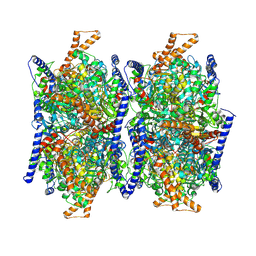 | |
8J78
 
 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state
To Be Published
|
|
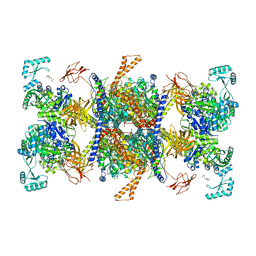
8J99
 
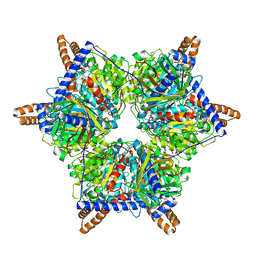 | | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, Methylcrotonoyl-CoA carboxylase subunit alpha, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state
To Be Published
|
|
8J7D
 
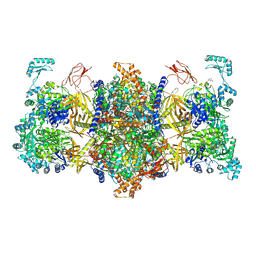 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state
To Be Published
|
|
8JAK
 
 | | Human MCC in MCCU state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Human MCC in BCCP-BCS state
To Be Published
|
|
8JAW
 
 | | Human MCC in MCCD state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Human MCC in BCCP-CTS state
To Be Published
|
|
8JXM
 
 | |
8JXL
 
 | |
8JXN
 
 | |
8K2V
 
 | |
6E5X
 
 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | Authors: | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-07-23 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
3DHB
 
 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DHC
 
 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
1JR6
 
 | |
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
2F3W
 
 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2QB3
 
 | | Structural Studies Reveal the Inactivation of E. coli L-Aspartate Aminotransferase by (s)-4,5-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 7.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2QA3
 
 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms (at pH6.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2R2D
 
 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | Authors: | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
2QBT
 
 | | Structural Studies Reveal The Inactivation of E. coli L-aspartate aminotransferase by (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 8.0) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-18 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
