6E5X
 
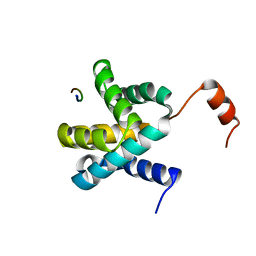 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | 分子名称: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | 著者 | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | 登録日 | 2018-07-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
1BNO
 
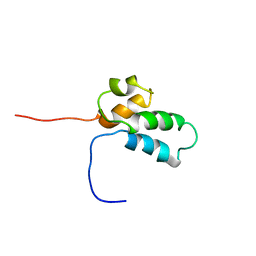 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | 登録日 | 1996-04-25 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BNP
 
 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | 分子名称: | DNA POLYMERASE BETA | | 著者 | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | 登録日 | 1996-04-25 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | 分子名称: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | 著者 | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
1ONB
 
 | |
1PFI
 
 | |
1JR6
 
 | |
1OVQ
 
 | |
1A4O
 
 | | 14-3-3 PROTEIN ZETA ISOFORM | | 分子名称: | 14-3-3 PROTEIN ZETA | | 著者 | Liu, D, Bienkowska, J, Petosa, C, Collier, R.J, Fu, H, Liddington, R.C. | | 登録日 | 1998-02-01 | | 公開日 | 1999-03-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the zeta isoform of the 14-3-3 protein.
Nature, 376, 1995
|
|
3DHB
 
 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | 分子名称: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | 著者 | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | 登録日 | 2008-06-17 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | 分子名称: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | 著者 | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | 登録日 | 2008-06-17 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DHC
 
 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | 分子名称: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | 著者 | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | 登録日 | 2008-06-17 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | 分子名称: | Thermonuclease | | 著者 | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1YYB
 
 | |
1RQ8
 
 | |
2A7M
 
 | | 1.6 Angstrom Resolution Structure of the Quorum-Quenching N-Acyl Homoserine Lactone Hydrolase of Bacillus thuringiensis | | 分子名称: | GLYCEROL, N-acyl homoserine lactone hydrolase, ZINC ION | | 著者 | Liu, D, Lepore, B.W, Petsko, G.A, Thomas, P.W, Stone, E.M, Fast, W, Ringe, D. | | 登録日 | 2005-07-05 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TBA
 
 | | SOLUTION STRUCTURE OF A TBP-TAFII230 COMPLEX: PROTEIN MIMICRY OF THE MINOR GROOVE SURFACE OF THE TATA BOX UNWOUND BY TBP, NMR, 25 STRUCTURES | | 分子名称: | TRANSCRIPTION INITIATION FACTOR IID 230K CHAIN, TRANSCRIPTION INITIATION FACTOR TFIID | | 著者 | Liu, D, Ishima, R, Tong, K.I, Bagby, S, Kokubo, T, Muhandiram, D.R, Kay, L.E, Nakatani, Y, Ikura, M. | | 登録日 | 1998-08-16 | | 公開日 | 1999-08-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP.
Cell(Cambridge,Mass.), 94, 1998
|
|
2F3W
 
 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | 分子名称: | Thermonuclease | | 著者 | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | 登録日 | 2005-11-22 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
2F3V
 
 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | 分子名称: | Thermonuclease | | 著者 | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | 登録日 | 2005-11-22 | | 公開日 | 2006-12-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
3EAZ
 
 | |
3PAA
 
 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 8.0 | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | 著者 | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | 登録日 | 2010-10-19 | | 公開日 | 2010-12-01 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
3PA9
 
 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 7.5 | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | 著者 | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | 登録日 | 2010-10-19 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
3EAC
 
 | |
7JRR
 
 | | Crystal structures of artificially designed homomeric RNA nanoarchitectures | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, RNA (50-MER) | | 著者 | Liu, D, Shao, Y, Piccirilli, J.A, Weizmann, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structures of artificially designed discrete RNA nanoarchitectures at near-atomic resolution.
Sci Adv, 7, 2021
|
|
4J51
 
 | |
