5O71
 
 | | Crystal structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Reverter, D, Liu, B. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | A quaternary tetramer assembly inhibits the deubiquitinating activity of USP25.
Nat Commun, 9, 2018
|
|
2WNM
 
 | | Solution structure of Gp2 | | Descriptor: | GENE 2 | | Authors: | Camara, B, Liu, M, Shadrinc, A, Liu, B, Simpson, P, Weinzierl, R, Severinovc, K, Cota, E, Matthews, S, Wigneshweraraj, S.R. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | T7 Phage Protein Gp2 Inhibits the Escherichia Coli RNA Polymerase by Antagonizing Stable DNA Strand Separation Near the Transcription Start Site.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8ZHP
 
 | | Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, Heavy chain of R1-32 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHF
 
 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHL
 
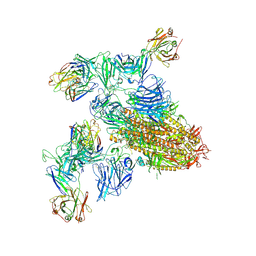 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHK
 
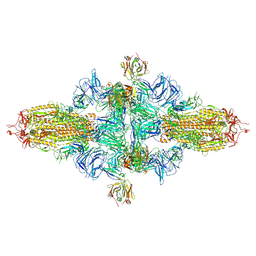 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHN
 
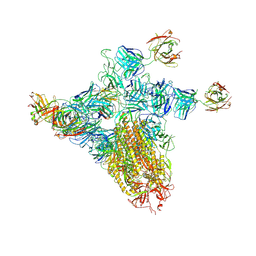 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs (one RBD rotated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHG
 
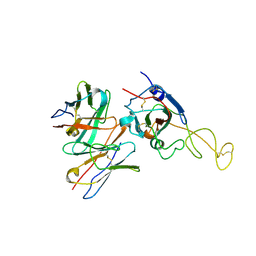 | | SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, focused refinement of RBD-Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, Light chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHI
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHD
 
 | | SARS-CoV-2 spike trimer (6P) in complex with two R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHJ
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C1 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHE
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-26 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHH
 
 | | SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.55 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
8ZHM
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants.
Nat Commun, 15, 2024
|
|
1HVS
 
 | | STRUCTURAL BASIS OF DRUG RESISTANCE FOR THE V82A MUTANT OF HIV-1 PROTEASE: BACKBONE FLEXIBILITY AND SUBSITE REPACKING | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Baldwin, E.T, Bhat, T.N, Liu, B, Pattabiraman, N, Erickson, J.W. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase.
Nat.Struct.Biol., 2, 1995
|
|
3UAF
 
 | | Crystal Structure of a TTR-52 mutant of C. elegans | | Descriptor: | TTR-52 | | Authors: | Kang, Y.Y, Zhao, D.F, Liang, H.H, Liu, B, Liu, Q.W, Wang, X.C, Liu, Y.F. | | Deposit date: | 2011-10-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of TTR-52 reveals the mechanism by which a bridging molecule mediates apoptotic cell engulfment
Genes Dev., 26, 2012
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
5H4D
 
 | | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 7-AMINO-4-METHYL-CHROMEN-2-ONE, ARG-HIS-LYS, ... | | Authors: | Zhang, S, Fu, L, Liu, J, Liu, B. | | Deposit date: | 2016-10-31 | | Release date: | 2017-11-08 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of hSIRT3 in complex with a specific agonist Amiodarone hydrochloride
To Be Published
|
|
7BOV
 
 | | The Structure of Bacillus subtilis glycosyltransferase,Bs-YjiC | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Zhao, C, Liu, B, Zhao, N.L, Luo, Y.Z, Bao, R. | | Deposit date: | 2020-03-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis.
Int.J.Biol.Macromol., 166, 2021
|
|
6GZ8
 
 | | First GerMN domain of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
5F5M
 
 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
1HDD
 
 | | CRYSTAL STRUCTURE OF AN ENGRAILED HOMEODOMAIN-DNA COMPLEX AT 2.8 ANGSTROMS RESOLUTION: A FRAMEWORK FOR UNDERSTANDING HOMEODOMAIN-DNA INTERACTIONS | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*TP*AP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*TP*AP*C P*CP*TP*AP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN) | | Authors: | Kissinger, C.R, Liu, B, Martin-Blanco, E, Kornberg, T.B, Pabo, C.O. | | Deposit date: | 1991-09-16 | | Release date: | 1992-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions.
Cell(Cambridge,Mass.), 63, 1990
|
|
7M8E
 
 | | E.coli RNAP-RapA elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, W, Liu, B. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation of Swi2/Snf2 ATPase RapA by RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
8ENR
 
 | |
