5IEL
 
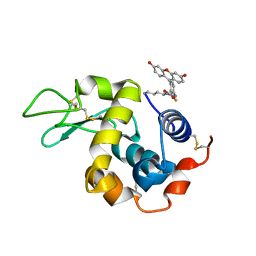 | | Structure of Lysozyme labeled with fluorescein isothiocyanate (FITC) at 1.15 Angstroms resolution | | Descriptor: | 2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-5-[(E)-(sulfanylmethylidene)amino]benzoic acid, Lysozyme C | | Authors: | Kachalova, G.S, Vlaskina, A.V, Popov, A.P, Simanovskaia, A.A, Krukova, M.V, Lipkin, A.V. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Lysozyme labeled with fluorescein isothiocyanate (FITC) at 1.15 Angstroms resolution
To Be Published
|
|
4RGZ
 
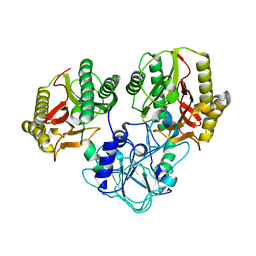 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5N9O
 
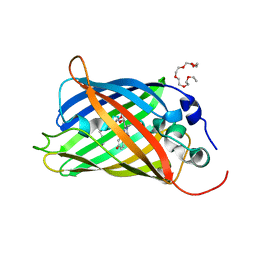 | | EGFP(enhanced green fluorescent protein) mutant - L232H | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Green fluorescent protein | | Authors: | Kachalova, G.S, Popov, A.P, Simanovskaya, A.A, Krukova, M.V, Lipkin, A.V. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure of EGFP(enhanced green fluorescent protein) mutant - L232H at 0.153 nm
To Be Published
|
|
1J5J
 
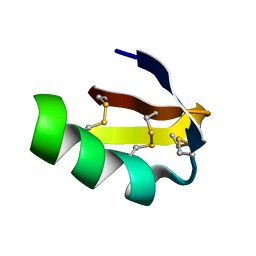 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
1LGL
 
 | | Solution structure of HERG-specific scorpion toxin BeKm-1 | | Descriptor: | BeKm-1 toxin | | Authors: | Korolokova, Y.V, Bocharov, E.V, Angelo, K, Maslennikov, I.V, Grinenko, O.V, Lipkin, A.V, Nosireva, E.D, Pluzhnikov, K.A, Olesen, S.-P, Arseniev, A.S, Grishin, E.V. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | New binding site on common molecular scaffold provides HERG channel specificity of scorpion toxin BeKm-1.
J.Biol.Chem., 277, 2002
|
|
4HX0
 
 | | Crystal structure of a putative nucleotidyltransferase (TM1012) from Thermotoga maritima at 1.87 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,4-BUTANEDIOL, Putative nucleotidyltransferase TM1012, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Lipkin, A.V, Popov, V.O, Kovalchuk, M.V, Shumilin, I.A, Minor, W, Shabalin, I.G, Golubev, A.M. | | Deposit date: | 2012-11-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of putative nucleotidyltransferase with two MPD molecules in the active site.
To be Published
|
|
4HGX
 
 | | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand | | Descriptor: | ACETATE ION, Xylose isomerase domain containing protein, ZINC ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Korzhenevskiy, D.A, Dorovatovsky, P.V, Rakitina, T.V, Lipkin, A.V, Shumilin, I.A, Minor, W, Popov, V.O. | | Deposit date: | 2012-10-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xylose isomerase domain containing protein (stm4435) from salmonella typhimurium lt2 with unknown ligand
To be Published
|
|
4NPO
 
 | | Crystal structure of protein with unknown function from Deinococcus radiodurans at P61 spacegroup | | Descriptor: | ACETATE ION, ACETYL GROUP, GLYCEROL, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Shabalin, I.G, Shumilin, I.A, Dorovatovsky, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of protein with unknown function at P61 spacegroup
To be Published
|
|
4NS4
 
 | | Crystal structure of cold-active estarase from Psychrobacter cryohalolentis K5T | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Boyko, K.M, Petrovskaya, L.E, Gorbacheva, M.A, Korgenevsky, D.A, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2013-11-28 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimentional structure of an esterse from Psychrobacter cryohalolentis K5T provides clues to unusual thermostability of a cold-active enzyme
To be Published
|
|
4NPA
 
 | | Scrystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korgenevsky, D.A, Dorovatovsky, P.V, Lipkin, A.V, Minor, W, Shumilin, I.A, Popov, V.O. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein with unknown function from Vibrio cholerae at P22121 spacegroup
To be Published
|
|
5L8Z
 
 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
4H05
 
 | | Crystal structure of aminoglycoside-3'-phosphotransferase of type VIII | | Descriptor: | Aminoglycoside-O-phosphotransferase VIII | | Authors: | Boyko, K.M, Gorbacheva, M.A, Danilenko, V.N, Alekseeva, M.G, Korzhenevskiy, D.A, Dorovatovskiy, P.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2012-09-07 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation.
Biochem.Biophys.Res.Commun., 477, 2016
|
|
