5F3R
 
 | |
6D99
 
 | |
6D9A
 
 | |
3MMJ
 
 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol hexakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-20 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
3MOZ
 
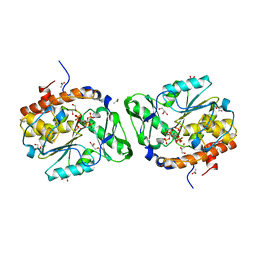 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,2,3,5,6)pentakisphosphate | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], ACETATE ION, CHLORIDE ION, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-23 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
4XAD
 
 | |
4XJJ
 
 | |
3GF4
 
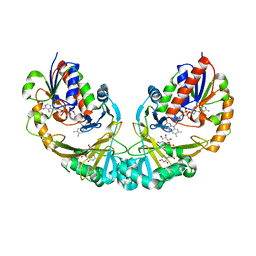 | | Structure of UDP-galactopyranose mutase bound to UDP-glucose | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Gruber, T.D, Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2009-02-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ligand binding and substrate discrimination by UDP-galactopyranose mutase.
J.Mol.Biol., 391, 2009
|
|
4KSI
 
 | | Crystal Structure Analysis of the Acidic Leucine Aminopeptidase of Tomato | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DuPrez, K.T, Scranton, M, Walling, L, Fan, L. | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of tomato wound-induced leucine aminopeptidase sheds light on substrate specificity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3INT
 
 | | Structure of UDP-galactopyranose mutase bound to UDP-galactose (reduced) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, Probable UDP-galactopyranose mutase, ... | | Authors: | Gruber, T.D, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin .
Biochemistry, 48, 2009
|
|
3INR
 
 | |
3F41
 
 | | Structure of the tandemly repeated protein tyrosine phosphatase like phytase from Mitsuokella multacida | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phytase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a multifunctional, tandemly repeated inositol polyphosphatase.
J.Mol.Biol., 392, 2009
|
|
7JN7
 
 | | Human DPP9-CARD8 complex | | Descriptor: | Caspase recruitment domain-containing protein 8, Dipeptidyl peptidase 9, [(2~{R})-1-[(2~{R})-2-azanyl-3-methyl-butanoyl]pyrrolidin-2-yl]boronic acid | | Authors: | Sharif, H, Hollingsworth, L.R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment.
Immunity, 54, 2021
|
|
7JKQ
 
 | | Human DPP9-CARD8 complex | | Descriptor: | Caspase recruitment domain-containing protein 8, Dipeptidyl peptidase 9 | | Authors: | Sharif, H, Hollingsworth, L.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dipeptidyl peptidase 9 sets a threshold for CARD8 inflammasome formation by sequestering its active C-terminal fragment.
Immunity, 54, 2021
|
|
3D1O
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 300 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
3D1H
 
 | |
4POG
 
 | | MCM-ssDNA co-crystal structure | | Descriptor: | 30-mer oligo(dT), Cell division control protein 21, ZINC ION | | Authors: | Froelich, C.A, Kang, S, Epling, L.B, Bell, S.P, Enemark, E.J. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Elife, 3, 2014
|
|
3D1Q
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 400 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
4POF
 
 | | PfMCM N-terminal domain without DNA | | Descriptor: | Cell division control protein 21, ZINC ION | | Authors: | Froelich, C.A, Kang, S, Epling, L.B, Bell, S.P, Enemark, E.J. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Elife, 3, 2014
|
|
6USC
 
 | | Structure of Human Intelectin-1 in complex with KO | | Descriptor: | CALCIUM ION, CHLORIDE ION, Intelectin-1, ... | | Authors: | Windsor, I.W, Isabella, C.R, Kosma, P, Raines, R.T, Kiessling, L.L. | | Deposit date: | 2019-10-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Stereoelectronic Effects Impact Glycan Recognition.
J.Am.Chem.Soc., 142, 2020
|
|
4QCJ
 
 | | Crystal Structure of OdhI from Corynebacterium glutamicum | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Labahn, J, Raasch, K, Eggeling, L, Bocola, M, Leitner, A, Bott, M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: Mutants and a model.
J.Biotechnol., 191, 2014
|
|
2QMT
 
 | | Crystal Polymorphism of Protein GB1 Examined by Solid-state NMR and X-ray Diffraction | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOPROPYL ALCOHOL, Immunoglobulin G-binding protein G, ... | | Authors: | Frericks Schmidt, H.L, Sperling, L.J, Gao, Y.G, Wylie, B.J, Boettcher, J.M, Wilson, S.R, Rienstra, C.M. | | Deposit date: | 2007-07-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal Polymorphism of Protein GB1 Examined by Solid-State NMR Spectroscopy and X-ray Diffraction.
J.Phys.Chem.B, 111, 2007
|
|
2FW0
 
 | | Apo Open Form of Glucose/Galactose Binding Protein | | Descriptor: | CALCIUM ION, CITRIC ACID, D-galactose-binding periplasmic protein, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2006-01-31 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures.
Protein Sci., 16, 2007
|
|
2FVY
 
 | | High Resolution Glucose Bound Crystal Structure of GGBP | | Descriptor: | ACETATE ION, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2006-01-31 | | Release date: | 2007-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Conformational changes of glucose/galactose-binding protein illuminated by open, unliganded, and ultra-high-resolution ligand-bound structures.
Protein Sci., 16, 2007
|
|
2QW1
 
 | | Glucose/galactose binding protein bound to 3-O-methyl D-glucose | | Descriptor: | 3-O-methyl-beta-D-glucopyranose, CALCIUM ION, D-galactose-binding periplasmic protein, ... | | Authors: | Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2007-08-09 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a periplasmic binding protein antagonist that prevents domain closure.
Acs Chem.Biol., 4, 2009
|
|
