5GCN
 
 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
7XC9
 
 | |
7XCF
 
 | |
7XCQ
 
 | |
6V6M
 
 | | Crystal structure of an inactive state of GMPPNP-bound RhoA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lin, Y, Zheng, Y. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure of an inactive conformation of GTP-bound RhoA GTPase.
Structure, 29, 2021
|
|
6V6U
 
 | |
8Z95
 
 | | Humanized anti-PEG h6.3 Fab in complex with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Heavy chain of Anti-PEG antibody h6-3 Fab fragment, ISOPROPYL ALCOHOL, ... | | Authors: | Lin, Y.C, Chang, C.Y, Su, Y.C. | | Deposit date: | 2024-04-22 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | pH-Responsive Polyethylene Glycol Engagers for Enhanced Brain Delivery of PEGylated Nanomedicine to Treat Glioblastoma.
Acs Nano, 19, 2025
|
|
8FPW
 
 | |
8FPX
 
 | |
5I10
 
 | | Crystal structure of spinosyn rhamnosyl 4'-O-methyltransferase spnh mutant T242Q from Saccharopolyspora Spinosa | | Descriptor: | MAGNESIUM ION, Probable O-methyltransferase | | Authors: | Lin, Y.-C, Huang, S.-P, Huang, B.-L, Chen, Y.-H, Chen, Y.-J, Chiu, H.-T. | | Deposit date: | 2016-02-05 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of spinosyn rhamnosyl 4'-O-methyltransferase spnh mutant T242Q from Saccharopolyspora Spinosa
To Be Published
|
|
6NIM
 
 | | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine | | Descriptor: | Bromodomain factor 2 protein, Bromosporine, SULFATE ION, ... | | Authors: | Lin, Y.H, Dong, A, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine
to be published
|
|
6NP7
 
 | | Crystal structure of Trypanosoma cruzi bromodomain BDF2 (TcCLB.506553.20) | | Descriptor: | Bromodomain factor 2 protein | | Authors: | Lin, Y.H, Dong, A, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Hui, R, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Trypanosoma cruzi bromodomain BDF2 (TcCLB.506553.20)
to be published
|
|
6NEZ
 
 | | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1 | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-[2-(4-propoxyphenyl)ethyl]-1H-benzimidazole, UNKNOWN ATOM OR ION, Uncharacterized protein | | Authors: | Lin, Y.H, Dong, A, Tempel, W, McAuley, J, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1
to be published
|
|
6MO0
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{3-[4-(furan-3-yl)phenyl]-5-[(piperidin-4-yl)methoxy]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J. Am. Chem. Soc., 141, 2019
|
|
6MO1
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 5-[4-(aminomethyl)phenyl]-6-[4-(furan-3-yl)phenyl]-N-[(piperidin-4-yl)methyl]pyrazin-2-amine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Hua, Y, Nie, S, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
6MO2
 
 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
9BB5
 
 | |
9BB7
 
 | |
9BB1
 
 | |
9BB6
 
 | |
9BB4
 
 | |
9BB2
 
 | |
9BB3
 
 | |
4GYS
 
 | |
9JKQ
 
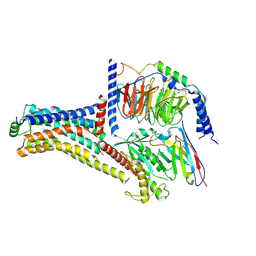 | | Cryo-EM structure of the METH-bound hTAAR1-Gs complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, Y, Wang, J, Shi, F. | | Deposit date: | 2024-09-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation.
J.Med.Chem., 67, 2024
|
|
